
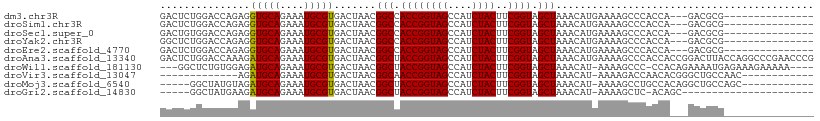
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,157,458 – 4,157,549 |
| Length | 91 |
| Max. P | 0.948151 |

| Location | 4,157,458 – 4,157,549 |
|---|---|
| Length | 91 |
| Sequences | 10 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 77.95 |
| Shannon entropy | 0.44435 |
| G+C content | 0.52450 |
| Mean single sequence MFE | -27.96 |
| Consensus MFE | -15.80 |
| Energy contribution | -15.55 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.54 |
| SVM RNA-class probability | 0.948151 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
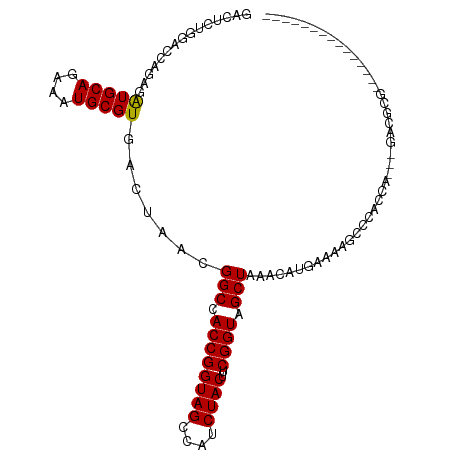
>dm3.chr3R 4157458 91 - 27905053 GACUCUGGACCAGAGGUGCAGAAAUGCGUGACUAACGGCCACCGGUAGCCAUCUACUUCGGUAGCUAAACAUGAAAAGCCCACCA---GACGCG--------------- (..((((((((.((((((((....)))(((.(((.(((...))).))).)))..)))))))).(((..........)))...)))---))..).--------------- ( -25.50, z-score = -1.07, R) >droSim1.chr3R 10408199 91 + 27517382 GACUCUGGACCAGAGGUGCAGAAAUGCGUGACUAACGGCCACCGGUAGCCAUCUACUUCGGUAGCUAAACAUGAAAAGCCCACCA---GACGCG--------------- (..((((((((.((((((((....)))(((.(((.(((...))).))).)))..)))))))).(((..........)))...)))---))..).--------------- ( -25.50, z-score = -1.07, R) >droSec1.super_0 3448782 91 + 21120651 GACUGUGGACCAGAGGUGCAGAAAUGCGUGACUAACGGCCACCGGUAGCCAUCUACUUCGGUAGCUAAACAUGAAAAGCCCACCA---GACGCG--------------- ..(((((((((.((((((((....)))(((.(((.(((...))).))).)))..)))))))).(((..........)))))).))---).....--------------- ( -24.70, z-score = -0.58, R) >droYak2.chr3R 8233611 91 - 28832112 GGCUCUGGACCAGAGGUGCAGAAAUGCGUGACUAACGGCCACCGGUAGCCAUCUACUUCGGUAGCUAAACAUGAAAAGCCCACCA---GACGCG--------------- .((((((((((.((((((((....)))(((.(((.(((...))).))).)))..)))))))).(((..........)))...)))---)).)).--------------- ( -28.60, z-score = -1.40, R) >droEre2.scaffold_4770 331504 91 + 17746568 GACUCUGGACCAGAGGUGCAGAAAUGCGUGACUAACGGCCACCGGUAGCCAUCUACUUCGGUAGCUAAACAUGAAAAGCCCACCA---GACGCG--------------- (..((((((((.((((((((....)))(((.(((.(((...))).))).)))..)))))))).(((..........)))...)))---))..).--------------- ( -25.50, z-score = -1.07, R) >droAna3.scaffold_13340 13256506 109 - 23697760 GACUCUGGACCAAAGAUGCAGAAAUGCGUGACUAACGGCUACCGGUAGCCAUCUACUUCGGUAGCUAAACAUGAAAAGCCCACCACCGGACUUACCAGGCCCGAACCCG ...((.((.((....(((((....))))).......((((((((((((....))))..)))))))).....((..(((.((......)).)))..)))))).))..... ( -29.80, z-score = -1.99, R) >droWil1.scaffold_181130 7564685 100 + 16660200 ---GGCUCUGUGGAGAUGCAGAAAUGCGUGACUAACGGCUACCGGUAGCCAUCUACUUCGGUAGCUAAACAU-AAAAGCCC-CCACAGAAAAUGAGAAAGAAAAA---- ---...(((((((..(((((....))))).......((((((((((((....))))..))))))))......-........-)))))))................---- ( -32.80, z-score = -4.09, R) >droVir3.scaffold_13047 15977460 83 - 19223366 -------------AGAUGCAGAAAUGCGUGACUAACGGCAACCGGUAGCCAUCUACUUCGGUAGCUAAACAU-AAAAGACCAACACGGGCUGCCAAC------------ -------------..(((((....))))).......(....).(((((((..((((....)))).....(..-....).........)))))))...------------ ( -24.50, z-score = -2.49, R) >droMoj3.scaffold_6540 19503992 91 - 34148556 -----GGCUAUGUAGAUGCAGAAAUGCGUGACUAACGGCUACCGGUAGCCAUCUACUUCGGUAGCUAAACAU-AAAAGCCUGCCACAGGCUGCCAGC------------ -----((((((((..(((((....))))).......((((((((((((....))))..))))))))..))))-)..((((((...)))))))))...------------ ( -37.20, z-score = -3.80, R) >droGri2.scaffold_14830 2230647 80 + 6267026 -----GGCUAUGAAGAUGCAGAAAUGCGUGACUAACGGCUACCGGUAGCCAUCUACUUCGGUAGCUAAACAU-AAAAGCUC-ACAGC---------------------- -----.(((.(((..(((((....))))).......((((((((((((....))))..))))))))......-......))-).)))---------------------- ( -25.50, z-score = -2.80, R) >consensus GACUCUGGACCAGAGAUGCAGAAAUGCGUGACUAACGGCCACCGGUAGCCAUCUACUUCGGUAGCUAAACAUGAAAAGCCCACCA___GACGCG_______________ ...............(((((....))))).......(((.((((((((....))))..)))).)))........................................... (-15.80 = -15.55 + -0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:58:09 2011