
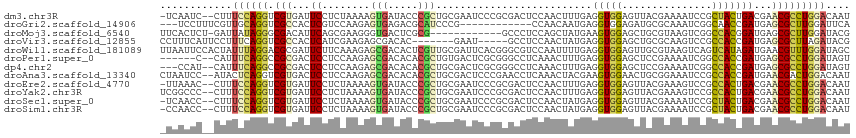
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,124,310 – 4,124,422 |
| Length | 112 |
| Max. P | 0.914037 |

| Location | 4,124,310 – 4,124,422 |
|---|---|
| Length | 112 |
| Sequences | 12 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 70.61 |
| Shannon entropy | 0.65946 |
| G+C content | 0.53805 |
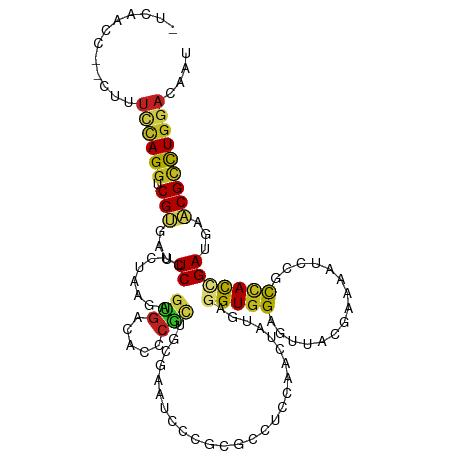
| Mean single sequence MFE | -32.72 |
| Consensus MFE | -14.12 |
| Energy contribution | -13.62 |
| Covariance contribution | -0.50 |
| Combinations/Pair | 1.63 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.914037 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 4124310 112 - 27905053 -UCAAUC--CUUUCCAGGUCGUGAUUCCUCUAAAAGUGAUACCCGCUGCGAAUCCCGCGACUCCAACUUUGAGGUGGAGUUACGAAAAUCCGCUACUGACGAACGCCUGGACAAU -......--...(((((((((((((((((((.(((((......(((..........)))......))))).))).)))))))))).....(((....).))....)))))).... ( -31.60, z-score = -2.25, R) >droGri2.scaffold_14906 12106474 100 + 14172833 ---UCCUUUCGUUGCAGGUCGCCACUCGUCCAAGAGUGAGACGCAUCCCG------------CCAACAAUGAGGUGGAGAUGCGCAAAUCGGCAACCGAUGAGCGCUUGGAUUCA ---(((...((((((..(((..(((((......))))).)))((((((((------------((........))))).)))))))..(((((...))))).))))...))).... ( -40.70, z-score = -3.01, R) >droMoj3.scaffold_6540 7558518 102 - 34148556 UUCACUCU-GAUUAUAGGGCGACAUUCAGCGAAGGGUGACUCGCG------------GCCCUCCAGCUAUGAAGUGGAGCUGCGUAAGUCGGCCACGGAUGAGCGCUUGGAUACG ........-.(((...(((((.(((((.((((..(....)))))(------------((((((((.((....)))))))..((....)).))))..)))))..))))).)))... ( -33.30, z-score = -0.06, R) >droVir3.scaffold_12855 9185579 103 + 10161210 CCUUUCAUUCCUUUCAGGUCGCCACUCAUCGAAGAGCGACAC-------GAAU-----GCCUCCAACUAUGAGGUGGAGCUGCGCAAGUCCGCCACCGAUGAGCGCUUAGAUACG .......((((.(((..(((((..((......)).)))))..-------))).-----(((((.......)))))))))..((((..(((.......)))..))))......... ( -25.20, z-score = 0.32, R) >droWil1.scaffold_181089 9566570 115 + 12369635 UUAAUUCCACUAUUUAGGACGCGAUUCUUCAAAGAGCGACACUCGUUGCGAUUCACGGGCGUCCAAUUUUGAGGUGGAGUUGCGUAAGUCAGUCAUAGAUGAACGUUUGGAUAGC ..(((((((((.....((((((((....)).....(((((....))))).........))))))........)))))))))((((..(((.......)))..))))......... ( -31.82, z-score = -1.13, R) >droPer1.super_0 6051233 107 + 11822988 ------C--CAUUUCAGGCCGCGACUCCUCCAAGAGCGACACACGCUGUGACUCGCGGGCCUCAAACUUUGAGGUGGAGCUCCGAAAAUCGGCCACCGAUGAGCGCCUGGAUAGU ------.--...((((((((((((.((.....(.((((.....)))).))).))))).((((((.....))))))...((((.....(((((...)))))))))))))))).... ( -38.80, z-score = -1.41, R) >dp4.chr2 14645978 110 - 30794189 ---CCAU--CAUUUCAGGCCGCGACUCCUCCAAGAGCGACACACGCUGCGACUCGCGGGCCUCAAACUUUGAGGUGGAGCUCCGAAAAUCGGCCACCGAUGAGCGCCUGGAUAGU ---....--...(((((((.((((((((.((...((((.....))))(((...)))))((((((.....)))))))))).)).....(((((...)))))..))))))))).... ( -39.20, z-score = -1.32, R) >droAna3.scaffold_13340 13221706 113 - 23697760 CUAAUCC--AUACUCAGGUCGUGACUCCUCCAAGAGCGACACACGCUGCGACUCCCGAACCUCAAACUACGAAGUGGAACUGCGGAAAUCCGCCACCGAUGAACGACUGGACAAU .......--....((.((((((...((.((((..((((.....)))).((.....)).................))))...((((....))))....))...)))))).)).... ( -23.40, z-score = 0.01, R) >droEre2.scaffold_4770 297291 112 + 17746568 -UUAAAC--CUUUCCAGGUCGUGAUUCCUCUAAAAGUGAUACCCGCUGCGAAUCCCGCGACUCCAACUUUGAGGUGGAGUUACGAAAGUCCGCCACUGACGAACGCCUGGACAAU -......--...(((((((((((((((((((.(((((......(((..........)))......))))).))).))))))))))..(((.......))).....)))))).... ( -33.20, z-score = -2.50, R) >droYak2.chr3R 8199523 113 - 28832112 UCGGCCC--CUUUCCAGGUCGUGAUUCCUCUAAAAGUGAUACCCGCUGCGAAUCCCGCGACUCCAACUUUGAGGUGGAGUUACGAAAGUCCGCCACUGACGAACGCCUGGACAAU .......--...(((((((((((((((((((.(((((......(((..........)))......))))).))).))))))))))..(((.......))).....)))))).... ( -33.20, z-score = -1.30, R) >droSec1.super_0 3415793 112 + 21120651 -UCAACC--CUUUCCAGGUCGUGAUUCCUCUAAAAGUGAUACCCGCUGCGAAUCCCGCGACUCCAACUAUGAGGUGGAGUUACGAAAAUCCGCUACUGACGAACGCCUGGACAAU -......--...((((((.(((...((.......((((.....))))(((.((..((..((((((.((....))))))))..))...)).)))....))...))))))))).... ( -31.10, z-score = -2.36, R) >droSim1.chr3R 10382601 112 + 27517382 -CCAACC--CUUUCCAGGUCGUGAUUCCUCUAAAAGUGAUACCCGCUGCGAAUCCCGCGACUCCAACUAUGAGGUGGAGUUACGAAAAUCCGCUACUGACGAACGCCUGGACAAU -......--...((((((.(((...((.......((((.....))))(((.((..((..((((((.((....))))))))..))...)).)))....))...))))))))).... ( -31.10, z-score = -2.50, R) >consensus _UCAACC__CUUUCCAGGUCGUGAUUCCUCUAAGAGUGACACCCGCUGCGAAUCCCGCGCCUCCAACUAUGAGGUGGAGUUACGAAAAUCCGCCACCGAUGAACGCCUGGACAAU ............((((((.(((...((........(((.....)))..........................(((((...............)))))))...))))))))).... (-14.12 = -13.62 + -0.50)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:58:07 2011