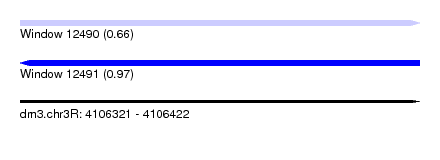
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,106,321 – 4,106,422 |
| Length | 101 |
| Max. P | 0.965653 |

| Location | 4,106,321 – 4,106,422 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 84.06 |
| Shannon entropy | 0.34194 |
| G+C content | 0.47171 |
| Mean single sequence MFE | -29.34 |
| Consensus MFE | -16.84 |
| Energy contribution | -15.97 |
| Covariance contribution | -0.87 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.37 |
| SVM RNA-class probability | 0.663447 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 4106321 101 + 27905053 AGUGUCGCUCU--UGUUAGCGACAAUUUACGGCUUUGAACGCUUCCGCACAAA-GCGCUGCCGUCAAUUUGUCAAUUACACAAAUUGUUCGGCUCCGUU-CGGUU------ ..(((((((..--....))))))).....((((((((...((....)).))))-))(..((((.((((((((.......))))))))..))))..)...-))...------ ( -32.30, z-score = -2.72, R) >droSim1.chr3R 10366894 101 - 27517382 AGUGUCGCUCU--UGUUAGCGACAAUUUACGGCUUUGAACGCUUCCGCACAAA-GCGCUGCCGUCAAUUUGUCAAUUACACAAAUUGUUCGGCUCCGUU-CGGUU------ ..(((((((..--....))))))).....((((((((...((....)).))))-))(..((((.((((((((.......))))))))..))))..)...-))...------ ( -32.30, z-score = -2.72, R) >droSec1.super_0 3398166 101 - 21120651 AGUGUCGCUCU--UGUUAGCGACAAUUUACGGCUUUGAACGCUUCCGCACAAA-GCGCUGCCGUCAAUUUGUCAAUUACACAAAUUGUUCGGCUCCGUU-CGGUU------ ..(((((((..--....))))))).....((((((((...((....)).))))-))(..((((.((((((((.......))))))))..))))..)...-))...------ ( -32.30, z-score = -2.72, R) >droEre2.scaffold_4770 277345 106 - 17746568 AGUGUCGCUCU--UGUUAGCGACAAUUUACGGCUUUGAACGCUUCCGCACAAA-GCGCUGCCGUCAAUUUGUCAAUUACACAAAUUGUUCGGCUCCGUUUCGGUUUUCC-- ..(((((((..--....))))))).....((((((((...((....)).))))-))(..((((.((((((((.......))))))))..))))..)....)).......-- ( -32.30, z-score = -2.87, R) >droYak2.chr3R 8181411 105 + 28832112 AGUGUCGCUCU--UGUUAGCGACAAUUUACGGCUUUGAACGCUUCCGCACAAA-GCGCUGCCGUCAAUUUGUCAAUUACACAAAUUGUUCGGCUCCGUU-CGCUUUUCC-- ..(((((((..--....)))))))....(((((((((...((....)).))))-))...((((.((((((((.......))))))))..))))..))).-.........-- ( -32.20, z-score = -3.12, R) >droAna3.scaffold_13340 13199924 107 + 23697760 AGUGUCGCUCU--UGUUAGCGACAAUUUACGGCUUUGAACGCUUCCGCACAAA-GCGCUGCCGUCAAUUUGUCAAUUACACAAAUUGUUUGGGUCCCCCUGGACUUGGAU- ..(((((((..--....)))))))....(((((......(((((.......))-)))..)))))((((((((.......))))))))((..(((((....)))))..)).- ( -35.10, z-score = -2.80, R) >dp4.chr2 14628931 99 + 30794189 CGCCUUGCUCU--UGUUAGCGACAAUUUACGGCUUCGAACGCUUCCGCACAAA-GCGUUGCCGUCAAUUUGUCAAUUACACAAAUUGUUUGGGUCCCAUCGU--------- .((((.(((..--....)))(((((.(((((((....(((((((.......))-)))))))))).)).))))).................))))........--------- ( -25.00, z-score = -1.13, R) >droPer1.super_0 6034257 99 - 11822988 CGCCUUGCUCU--UGUUAGCGACAAUUUACGGCUUCGAACGCUUCCGCACAAA-GCGUUGCCGUCAAUUUGUCAAUUACACAAAUUGUUUGGGUCCCAUCGU--------- .((((.(((..--....)))(((((.(((((((....(((((((.......))-)))))))))).)).))))).................))))........--------- ( -25.00, z-score = -1.13, R) >droWil1.scaffold_181089 9551479 105 - 12369635 UGUGUCGCUCU--UGUUAGCGACAAUUUACGGCUUUGAACGCUUCCGCACAAA-GCGUUGCCGUCAAUUUGUCAAUUACACAAAUUGUUUAGGUCUCCCCGACAGAAU--- ..(((((((..--....)))))))....(((((....(((((((.......))-))))))))))((((((((.......)))))))).....(((.....))).....--- ( -30.10, z-score = -2.57, R) >droVir3.scaffold_12855 9167114 108 - 10161210 GAUCGCGCUCU--UGUUAGCGACAAUUUACGGCUUUGAACGCUUCCGCACAAU-GCGUCGCCGUCAAUUUGUCAAUUACACAAAUUGUUUAGGUCUUCUCUCGCCACGAUG .((((((((..--....)))).........(((...((.(((...........-)))))(((..((((((((.......))))))))....)))........))).)))). ( -24.80, z-score = -0.57, R) >droMoj3.scaffold_6540 7542024 108 + 34148556 GAUCGCGCUCU--UGUUAGCGACAAUUUACGGCUUUGAACGCUUCCGCACAAU-GCGUCGCCGUCAAUUUGUCAAUUACACAAAUUGUUUAGCUCUCGACUCGCCACGACA ....(((...(--((..((((((((.(((((((...((.(((...........-)))))))))).)).)))))....(((.....)))...)))..)))..)))....... ( -24.20, z-score = -0.13, R) >droGri2.scaffold_14906 12088649 111 - 14172833 GAUCGAGCUCUCUUGUUAGCGACAAUUUACGGCUUUGAACGCUUCCGCACAAAAGCGUCGCCGUCAAUUUGUCAAUUACACAAAUUGUUUAGGUCUCGGCUCACCACGAUA ....(((((..((((..((((((((.(((((((...((.(((((........)))))))))))).)).)))))((((.....)))))))))))....)))))......... ( -26.50, z-score = -0.98, R) >consensus AGUGUCGCUCU__UGUUAGCGACAAUUUACGGCUUUGAACGCUUCCGCACAAA_GCGCUGCCGUCAAUUUGUCAAUUACACAAAUUGUUUGGCUCCCUUCCGGUU______ .....((((........)))).........((..((((((((...(((......)))..))....(((((((.......)))))))))))))..))............... (-16.84 = -15.97 + -0.87)

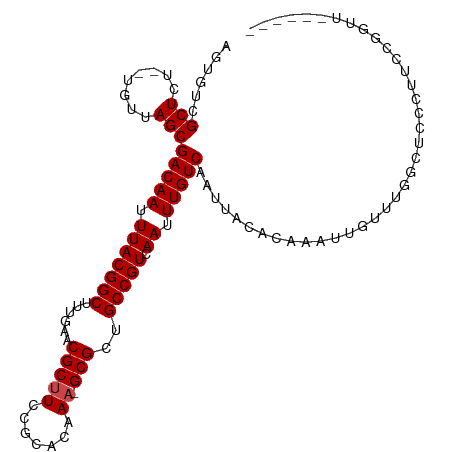
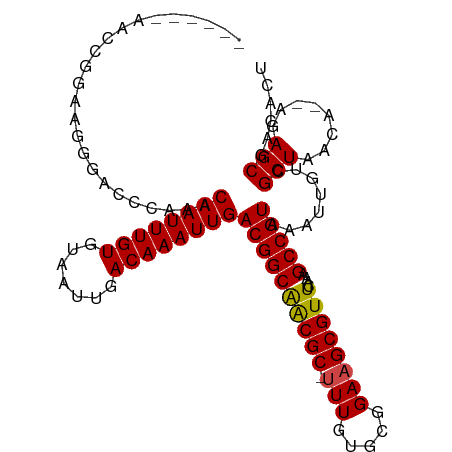
| Location | 4,106,321 – 4,106,422 |
|---|---|
| Length | 101 |
| Sequences | 12 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 84.06 |
| Shannon entropy | 0.34194 |
| G+C content | 0.47171 |
| Mean single sequence MFE | -34.27 |
| Consensus MFE | -22.21 |
| Energy contribution | -21.94 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.41 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.965653 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 4106321 101 - 27905053 ------AACCG-AACGGAGCCGAACAAUUUGUGUAAUUGACAAAUUGACGGCAGCGC-UUUGUGCGGAAGCGUUCAAAGCCGUAAAUUGUCGCUAACA--AGAGCGACACU ------.....-.(((..((((..((((((((.......)))))))).))))...((-(((((((....)))..)))))))))....(((((((....--..))))))).. ( -37.30, z-score = -3.69, R) >droSim1.chr3R 10366894 101 + 27517382 ------AACCG-AACGGAGCCGAACAAUUUGUGUAAUUGACAAAUUGACGGCAGCGC-UUUGUGCGGAAGCGUUCAAAGCCGUAAAUUGUCGCUAACA--AGAGCGACACU ------.....-.(((..((((..((((((((.......)))))))).))))...((-(((((((....)))..)))))))))....(((((((....--..))))))).. ( -37.30, z-score = -3.69, R) >droSec1.super_0 3398166 101 + 21120651 ------AACCG-AACGGAGCCGAACAAUUUGUGUAAUUGACAAAUUGACGGCAGCGC-UUUGUGCGGAAGCGUUCAAAGCCGUAAAUUGUCGCUAACA--AGAGCGACACU ------.....-.(((..((((..((((((((.......)))))))).))))...((-(((((((....)))..)))))))))....(((((((....--..))))))).. ( -37.30, z-score = -3.69, R) >droEre2.scaffold_4770 277345 106 + 17746568 --GGAAAACCGAAACGGAGCCGAACAAUUUGUGUAAUUGACAAAUUGACGGCAGCGC-UUUGUGCGGAAGCGUUCAAAGCCGUAAAUUGUCGCUAACA--AGAGCGACACU --((....))...(((..((((..((((((((.......)))))))).))))...((-(((((((....)))..)))))))))....(((((((....--..))))))).. ( -37.80, z-score = -3.67, R) >droYak2.chr3R 8181411 105 - 28832112 --GGAAAAGCG-AACGGAGCCGAACAAUUUGUGUAAUUGACAAAUUGACGGCAGCGC-UUUGUGCGGAAGCGUUCAAAGCCGUAAAUUGUCGCUAACA--AGAGCGACACU --......(((-......((((..((((((((.......)))))))).))))...((-(((((((....)))..)))))))))....(((((((....--..))))))).. ( -37.40, z-score = -3.24, R) >droAna3.scaffold_13340 13199924 107 - 23697760 -AUCCAAGUCCAGGGGGACCCAAACAAUUUGUGUAAUUGACAAAUUGACGGCAGCGC-UUUGUGCGGAAGCGUUCAAAGCCGUAAAUUGUCGCUAACA--AGAGCGACACU -......((((....)))).....((((((((.......))))))))((((((((((-(((.....))))))))....)))))....(((((((....--..))))))).. ( -37.00, z-score = -3.01, R) >dp4.chr2 14628931 99 - 30794189 ---------ACGAUGGGACCCAAACAAUUUGUGUAAUUGACAAAUUGACGGCAACGC-UUUGUGCGGAAGCGUUCGAAGCCGUAAAUUGUCGCUAACA--AGAGCAAGGCG ---------..((..(........((((((((.......))))))))((((((((((-(((.....))))))))....)))))...)..))(((....--..)))...... ( -27.90, z-score = -0.88, R) >droPer1.super_0 6034257 99 + 11822988 ---------ACGAUGGGACCCAAACAAUUUGUGUAAUUGACAAAUUGACGGCAACGC-UUUGUGCGGAAGCGUUCGAAGCCGUAAAUUGUCGCUAACA--AGAGCAAGGCG ---------..((..(........((((((((.......))))))))((((((((((-(((.....))))))))....)))))...)..))(((....--..)))...... ( -27.90, z-score = -0.88, R) >droWil1.scaffold_181089 9551479 105 + 12369635 ---AUUCUGUCGGGGAGACCUAAACAAUUUGUGUAAUUGACAAAUUGACGGCAACGC-UUUGUGCGGAAGCGUUCAAAGCCGUAAAUUGUCGCUAACA--AGAGCGACACA ---....(((((((....))....((((((((.......)))))))).)))))((((-(((((((....)))..))))).)))....(((((((....--..))))))).. ( -38.80, z-score = -4.14, R) >droVir3.scaffold_12855 9167114 108 + 10161210 CAUCGUGGCGAGAGAAGACCUAAACAAUUUGUGUAAUUGACAAAUUGACGGCGACGC-AUUGUGCGGAAGCGUUCAAAGCCGUAAAUUGUCGCUAACA--AGAGCGCGAUC .((((((((((.............((((((((.......))))))))(((((.....-....(((....)))......)))))...)))))(((....--..)))))))). ( -30.16, z-score = -0.82, R) >droMoj3.scaffold_6540 7542024 108 - 34148556 UGUCGUGGCGAGUCGAGAGCUAAACAAUUUGUGUAAUUGACAAAUUGACGGCGACGC-AUUGUGCGGAAGCGUUCAAAGCCGUAAAUUGUCGCUAACA--AGAGCGCGAUC .((((((((((.............((((((((.......))))))))(((((.....-....(((....)))......)))))...)))))(((....--..)))))))). ( -30.36, z-score = 0.17, R) >droGri2.scaffold_14906 12088649 111 + 14172833 UAUCGUGGUGAGCCGAGACCUAAACAAUUUGUGUAAUUGACAAAUUGACGGCGACGCUUUUGUGCGGAAGCGUUCAAAGCCGUAAAUUGUCGCUAACAAGAGAGCUCGAUC .(((((((....))).(((.....((((((((.......))))))))((((((((((((((....)))))))))....))))).....)))(((........))).)))). ( -32.00, z-score = -1.35, R) >consensus ______AACCGGAAGGGACCCAAACAAUUUGUGUAAUUGACAAAUUGACGGCAACGC_UUUGUGCGGAAGCGUUCAAAGCCGUAAAUUGUCGCUAACA__AGAGCGACACU ........................((((((((.......))))))))((((((((((.(((....))).)))))....)))))........(((........)))...... (-22.21 = -21.94 + -0.27)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:58:06 2011