
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,105,487 – 4,105,561 |
| Length | 74 |
| Max. P | 0.684243 |

| Location | 4,105,487 – 4,105,561 |
|---|---|
| Length | 74 |
| Sequences | 12 |
| Columns | 77 |
| Reading direction | forward |
| Mean pairwise identity | 76.67 |
| Shannon entropy | 0.50836 |
| G+C content | 0.48452 |
| Mean single sequence MFE | -15.99 |
| Consensus MFE | -11.37 |
| Energy contribution | -11.46 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.43 |
| Mean z-score | -0.68 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.684243 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 4105487 74 + 27905053 GCCCCGAAAAUCAAGCGCCACU-CACA--AAUCGUUUAAUACGAAGUUCGAUCCCCGAUCGUUGUUGGCGGAUAAAC ...............(((((..-.(((--(.((((.....))))....(((((...))))))))))))))....... ( -15.70, z-score = -0.94, R) >droSim1.chr3R 10366076 74 - 27517382 GUCCCGAAAAUCAAGCGCCACU-CACA--AAUCGUUUAAUGCGAAGUUCGAUCCCCGAUCGUUGUUGGCGGAUAACC ((((.(.....)....((((..-.(((--(.((((.....))))....(((((...))))))))))))))))).... ( -16.40, z-score = -0.71, R) >droSec1.super_0 3397344 74 - 21120651 GUCCCGAAAAUCAAGCGCCACU-CACA--AAUCGUUUAAUGCGAAGUUCGAUCCCCGAUCGUUGUUGGCGGAUAACC ((((.(.....)....((((..-.(((--(.((((.....))))....(((((...))))))))))))))))).... ( -16.40, z-score = -0.71, R) >droEre2.scaffold_4770 276528 74 - 17746568 GUCCCAAAAACCAAGCGCCACU-CACA--AAUCGCUUAAUGCGAAGUUCGAUCCCCGAUCGUUGUUGGCGGAUAACC ...............(((((..-.(((--(.((((.....))))....(((((...))))))))))))))....... ( -18.10, z-score = -1.93, R) >droYak2.chr3R 8180496 74 + 28832112 GUCCCGAAAACCAAGCGCCACU-AACA--AAUCGUUUAAUGCGAAGUUCGAUCCCCGAUCGUUGUUGGCGGAUAACC ...............(((((.(-(((.--..((((.....)))).....((((...)))))))).)))))....... ( -17.80, z-score = -1.48, R) >droAna3.scaffold_13340 13199091 74 + 23697760 GACCCGAAAAUCAAGCGCCACU-CACA--AAUCGUUUAAGUCGAAGUUCGAUCCUCGAUCGUUGUUGGUGGAUAAAC .................(((((-.(((--(((((.....((((.....))))...))))..)))).)))))...... ( -16.00, z-score = -0.58, R) >dp4.chr2 14628245 77 + 30794189 GUCCCGAAAAUCAAGCGCCGCCACUCACAAAUCGUUUAAGGCGAAGUUCGAUCGAUGAUCGUUGCUGGCGGAUAACC .................(((((.........((((.....))))(((.(((((...)))))..))))))))...... ( -19.40, z-score = -0.30, R) >droPer1.super_0 6033501 77 - 11822988 GUCCCGAAAAUCAAGCGCCGCCACUCACAAAUCGUUUAAGGCGAAGUUCGAUCGGUGAUCGUUGCUGGCGGAUAACC .................(((((.........((((.....))))(((.(((((...)))))..))))))))...... ( -19.40, z-score = 0.09, R) >droWil1.scaffold_181089 9550846 63 - 12369635 GUUCCAAAAAACAAAGAGGGCAAAAAACAAAUCGUUUAAAGUGAACUUCGAUCAACGAUCGUU-------------- ...............(((..((..((((.....))))....))..)))(((((...)))))..-------------- ( -7.20, z-score = 0.05, R) >droGri2.scaffold_14906 12088030 64 - 14172833 ------------GAGCGCCACU-CACACAAAUCGUUUAAAGCGAACAUCGAUGGAUGAUCGCUGUUGGUGGAUAACC ------------.....(((((-.((((.....))....(((((.((((....)))).))))))).)))))...... ( -17.90, z-score = -1.77, R) >droMoj3.scaffold_6540 7541433 65 + 34148556 ---------AUCGAGCGCCACU-CACA--AAUCGUUUAAAGCGAAGUUCGAUUGAUGAUCGUUGCUGGCGGAUAACC ---------......((((...-....--..((((.....))))(((.(((((...)))))..)))))))....... ( -14.70, z-score = -0.02, R) >droVir3.scaffold_12855 9166538 65 - 10161210 ---------AUCGAGCACCACU-CACA--AAUCGUUUAAAGCGAACUCCGAUUGAUGAUCGUGGCUGGCGGAUAACC ---------(((..((.(((((-((..--(((((..............)))))..)))..))))...)).))).... ( -12.94, z-score = 0.11, R) >consensus GUCCCGAAAAUCAAGCGCCACU_CACA__AAUCGUUUAAUGCGAAGUUCGAUCCACGAUCGUUGUUGGCGGAUAACC ...............(((((...........((((.....))))....(((((...)))))....)))))....... (-11.37 = -11.46 + 0.09)

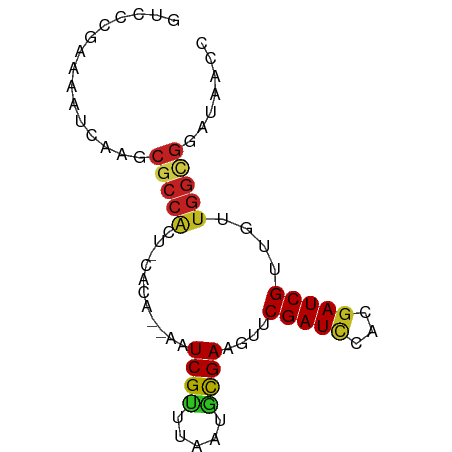
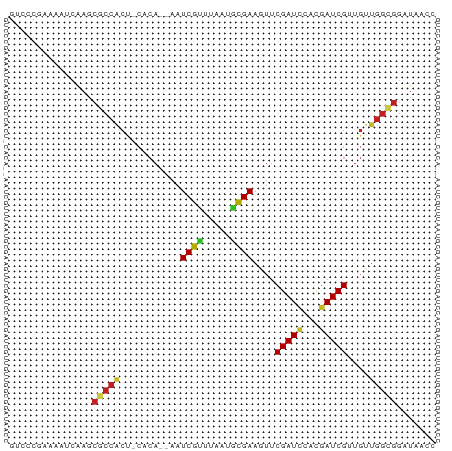
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:58:04 2011