| Sequence ID | dm3.chr3R |
|---|---|
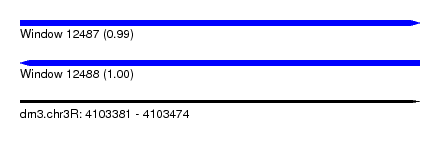
| Location | 4,103,381 – 4,103,474 |
| Length | 93 |
| Max. P | 0.995330 |

| Location | 4,103,381 – 4,103,474 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 86.18 |
| Shannon entropy | 0.26918 |
| G+C content | 0.39712 |
| Mean single sequence MFE | -24.22 |
| Consensus MFE | -19.50 |
| Energy contribution | -20.20 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.989571 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
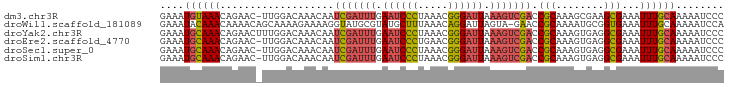
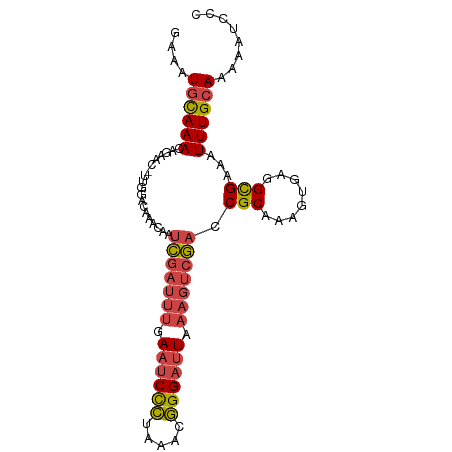
>dm3.chr3R 4103381 93 + 27905053 GGGAUUUUUGCAAAUUUCGCUUCGCUUUGCGGUCGACUUUAAUCCCGUUUAGGGAUUCAAAUCGAUUGUUUGUCCAA-GUUCUGUUUACAUUUC (((((((..((((((..(((........)))(((((.((.((((((.....)))))).)).))))).))))))..))-)))))........... ( -25.80, z-score = -3.16, R) >droWil1.scaffold_181089 9548320 93 - 12369635 UGGAUUUUUGCAAAUUUCACCGCAUUUUGCGGUUC-UACUAAUCCUGUUUAAAGCAUACGCAUACCUUUUCUUUUGCUGUUUUGUUUGUAUUUC ........(((((((...(((((.....)))))..-..............((((((...(((............)))))))))))))))).... ( -15.30, z-score = -1.17, R) >droYak2.chr3R 8178324 94 + 28832112 GGGAUUUUUGCAAAUUUCGCCUCACUUUGCGGUCGACUUUAAUCCCGUUUAGGGAUUCAAAUCGAUUGUUUGUCCAAAGUUCUGUUUGCAUUUC ........(((((((...((...((...((((((((.((.((((((.....)))))).)).))))))))..)).....))...))))))).... ( -25.60, z-score = -2.66, R) >droEre2.scaffold_4770 274339 93 - 17746568 GGGAUUUUUGCAAAUUUCGCCUCACUUUGCGGUCGACUUUAAUCCCGUUCAGGGAUUCAAAUCGAUUGUUUGUCCAA-GUUCUGUUUGCAUUUC ........(((((((...((...((...((((((((.((.((((((.....)))))).)).))))))))..))....-))...))))))).... ( -26.20, z-score = -2.86, R) >droSec1.super_0 3395202 93 - 21120651 GGGAUUUUUGCAAAUUUCGCCUCACUUUGCGGUCGACUUUAAUCCCGUUUAGGGAUUCAAAUCGAUUGUUUGUCCAA-GUUCUGUUUGCAUUUC ........(((((((...((...((...((((((((.((.((((((.....)))))).)).))))))))..))....-))...))))))).... ( -26.20, z-score = -3.01, R) >droSim1.chr3R 10363923 93 - 27517382 GGGAUUUUUGCAAAUUUCGCCUCACUUUGCGGUCGACUUUAAUCCCGUUUAGGGAUUCAAAUCGAUUGUUUGUCCAA-GUUCUGUUUGCAUUUC ........(((((((...((...((...((((((((.((.((((((.....)))))).)).))))))))..))....-))...))))))).... ( -26.20, z-score = -3.01, R) >consensus GGGAUUUUUGCAAAUUUCGCCUCACUUUGCGGUCGACUUUAAUCCCGUUUAGGGAUUCAAAUCGAUUGUUUGUCCAA_GUUCUGUUUGCAUUUC ........(((((((...((........((((((((.((.((((((.....)))))).)).)))))))).........))...))))))).... (-19.50 = -20.20 + 0.70)

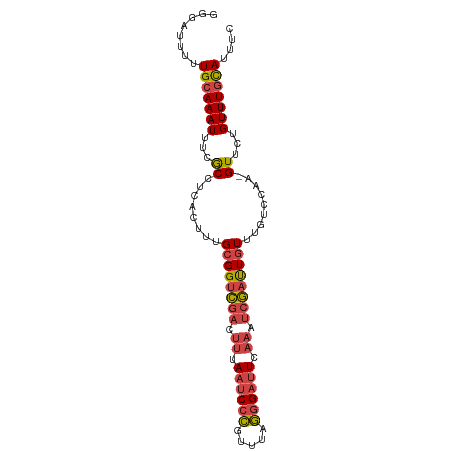
| Location | 4,103,381 – 4,103,474 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 86.18 |
| Shannon entropy | 0.26918 |
| G+C content | 0.39712 |
| Mean single sequence MFE | -22.85 |
| Consensus MFE | -18.70 |
| Energy contribution | -19.23 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.95 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.79 |
| SVM RNA-class probability | 0.995330 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 4103381 93 - 27905053 GAAAUGUAAACAGAAC-UUGGACAAACAAUCGAUUUGAAUCCCUAAACGGGAUUAAAGUCGACCGCAAAGCGAAGCGAAAUUUGCAAAAAUCCC ................-..(((.......(((((((.((((((.....)))))).)))))))..(((((.((...))...))))).....))). ( -22.50, z-score = -3.21, R) >droWil1.scaffold_181089 9548320 93 + 12369635 GAAAUACAAACAAAACAGCAAAAGAAAAGGUAUGCGUAUGCUUUAAACAGGAUUAGUA-GAACCGCAAAAUGCGGUGAAAUUUGCAAAAAUCCA ..........................(((((((....))))))).....(((((.(((-(((((((.....)))))....)))))...))))). ( -17.80, z-score = -1.92, R) >droYak2.chr3R 8178324 94 - 28832112 GAAAUGCAAACAGAACUUUGGACAAACAAUCGAUUUGAAUCCCUAAACGGGAUUAAAGUCGACCGCAAAGUGAGGCGAAAUUUGCAAAAAUCCC ....((((((...................(((((((.((((((.....)))))).))))))).(((........)))...))))))........ ( -24.20, z-score = -3.09, R) >droEre2.scaffold_4770 274339 93 + 17746568 GAAAUGCAAACAGAAC-UUGGACAAACAAUCGAUUUGAAUCCCUGAACGGGAUUAAAGUCGACCGCAAAGUGAGGCGAAAUUUGCAAAAAUCCC ....((((((......-............(((((((.((((((.....)))))).))))))).(((........)))...))))))........ ( -24.20, z-score = -2.94, R) >droSec1.super_0 3395202 93 + 21120651 GAAAUGCAAACAGAAC-UUGGACAAACAAUCGAUUUGAAUCCCUAAACGGGAUUAAAGUCGACCGCAAAGUGAGGCGAAAUUUGCAAAAAUCCC ....((((((......-............(((((((.((((((.....)))))).))))))).(((........)))...))))))........ ( -24.20, z-score = -3.26, R) >droSim1.chr3R 10363923 93 + 27517382 GAAAUGCAAACAGAAC-UUGGACAAACAAUCGAUUUGAAUCCCUAAACGGGAUUAAAGUCGACCGCAAAGUGAGGCGAAAUUUGCAAAAAUCCC ....((((((......-............(((((((.((((((.....)))))).))))))).(((........)))...))))))........ ( -24.20, z-score = -3.26, R) >consensus GAAAUGCAAACAGAAC_UUGGACAAACAAUCGAUUUGAAUCCCUAAACGGGAUUAAAGUCGACCGCAAAGUGAGGCGAAAUUUGCAAAAAUCCC ....((((((...................(((((((.((((((.....)))))).))))))).(((........)))...))))))........ (-18.70 = -19.23 + 0.53)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:58:04 2011