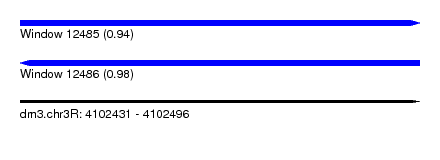
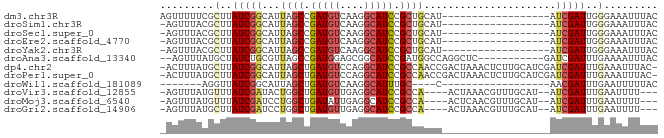
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,102,431 – 4,102,496 |
| Length | 65 |
| Max. P | 0.981203 |

| Location | 4,102,431 – 4,102,496 |
|---|---|
| Length | 65 |
| Sequences | 12 |
| Columns | 83 |
| Reading direction | forward |
| Mean pairwise identity | 65.40 |
| Shannon entropy | 0.67390 |
| G+C content | 0.42901 |
| Mean single sequence MFE | -15.63 |
| Consensus MFE | -7.40 |
| Energy contribution | -7.20 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.943880 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
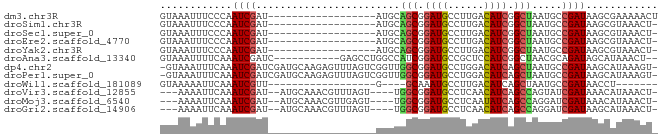
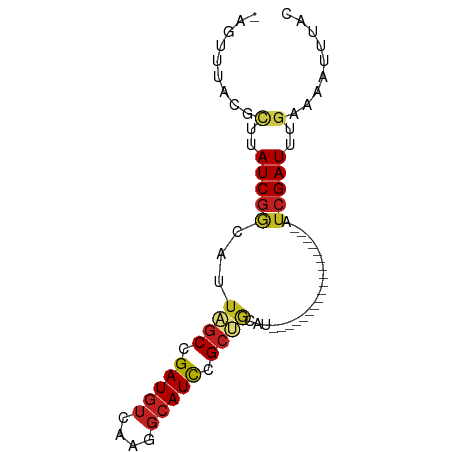
>dm3.chr3R 4102431 65 + 27905053 GUAAAUUUCCCAAUCGAU------------------AUGCAGCGGAUGCCUUGACAUCGGCUAAUGCCGAUAAGCGAAAAACU .....(((((..((((.(------------------((..(((.((((......)))).))).))).))))..).)))).... ( -15.40, z-score = -1.53, R) >droSim1.chr3R 10362975 64 - 27517382 GUAAAUUUCCCAAUCGAU------------------AUGCAGCGGAUGCCUUGACAUCGGCUAAUGCCGAUAAGCGUAAACU- .................(------------------((((.((....))......((((((....))))))..)))))....- ( -14.80, z-score = -1.08, R) >droSec1.super_0 3394254 64 - 21120651 GUAAAUUUCCCAAUCGAU------------------AUGCAGCGGAUGCCUUGACAUCGGCUAAUGCCGAUAAGCGUAAACU- .................(------------------((((.((....))......((((((....))))))..)))))....- ( -14.80, z-score = -1.08, R) >droEre2.scaffold_4770 273379 64 - 17746568 GUAAAUUUCCCAAUCGAU------------------AUGCAGCGGAUGCCUUGACAUCGGCUAAUGCCGAUAAGCGUAAACU- .................(------------------((((.((....))......((((((....))))))..)))))....- ( -14.80, z-score = -1.08, R) >droYak2.chr3R 8177361 64 + 28832112 GUAAAUUUCCCAAUCGAU------------------AUGCAGCGGAUGCCUUGACAUCGGCUAAUGCCGAUAAGCGUAAACU- .................(------------------((((.((....))......((((((....))))))..)))))....- ( -14.80, z-score = -1.08, R) >droAna3.scaffold_13340 13195611 70 + 23697760 GUAAAUUUUCAAAUCGAUC-----------GAGCCUGGCCAUCGGAUGCCGCUCCAUCGGCUAACGCAGAUAGCAUAAACU-- ((............((((.-----------((((..(((........))))))).))))((....)).....)).......-- ( -16.20, z-score = -0.64, R) >dp4.chr2 14624806 81 + 30794189 -GUAAAUUUCAAAUCGAUCGAUGCAAGAGUUUAGUCGGUUGGCGGAUGCCUGGACAUCAGCUAAUGCCGAUAAGCAUAAAGU- -.(((((((...(((....)))....)))))))((((((((((.((((......)))).))))..))))))...........- ( -19.90, z-score = -0.96, R) >droPer1.super_0 6030063 81 - 11822988 -GUAAAUUUCAAAUCGAUCGAUGCAAGAGUUUAGUCGGUUGGCGGAUGCCUGGACAUCAGCUAAUGCCGAUAAGCAUAAAGU- -.(((((((...(((....)))....)))))))((((((((((.((((......)))).))))..))))))...........- ( -19.90, z-score = -0.96, R) >droWil1.scaffold_181089 9547397 54 - 12369635 GUAAAAAUUCAAAUCGUU------------------G----GCAAAUGCCUUGACAUCAGCUAAUGCCGAUAACCU------- ...............(((------------------(----(((...((..((....))))...))))))).....------- ( -10.10, z-score = -1.87, R) >droVir3.scaffold_12855 9162881 73 - 10161210 ---AAAAUUCAAAUCGAU--AUGCAAACGUUUAGU----UGGCGGAUGCCUCAACAUCAGCCAGUAUCGAUAAACAUAAACU- ---.........((((((--((...(((.....))----)(((.((((......)))).))).))))))))...........- ( -18.00, z-score = -3.35, R) >droMoj3.scaffold_6540 7537580 73 + 34148556 ---AAAAUUCAAAUCGAU--AUGCAAACGUUGAGU----UGGCGGAUGCCUCAAUAUCAGCCAGGAUCGAUAAACAUAAACU- ---..............(--(((.....(((((.(----((((.((((......)))).)))))..)))))...))))....- ( -13.50, z-score = -0.79, R) >droGri2.scaffold_14906 12083687 73 - 14172833 ---AAAAUUCAAAUCGAU--AUGCAAACGUUUAGU----UGGCGGAUGCCUCAACAUCAGCCAGGAUCGAUAAGCAUAAACU- ---.........((((((--.....(((.....))----)(((.((((......)))).)))...))))))...........- ( -15.40, z-score = -1.46, R) >consensus GUAAAAUUUCAAAUCGAU__________________AUGCAGCGGAUGCCUUGACAUCAGCUAAUGCCGAUAAGCAUAAACU_ ........................................(((.((((......)))).)))..................... ( -7.40 = -7.20 + -0.20)
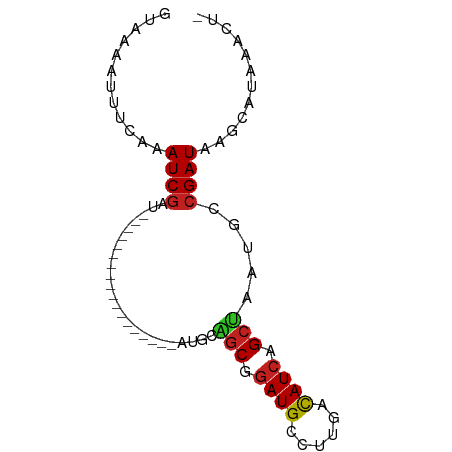
| Location | 4,102,431 – 4,102,496 |
|---|---|
| Length | 65 |
| Sequences | 12 |
| Columns | 83 |
| Reading direction | reverse |
| Mean pairwise identity | 65.40 |
| Shannon entropy | 0.67390 |
| G+C content | 0.42901 |
| Mean single sequence MFE | -17.01 |
| Consensus MFE | -8.50 |
| Energy contribution | -8.55 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.981203 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 4102431 65 - 27905053 AGUUUUUCGCUUAUCGGCAUUAGCCGAUGUCAAGGCAUCCGCUGCAU------------------AUCGAUUGGGAAAUUUAC ....((((.((.((((..((((((.(((((....))))).)))).))------------------..)))).))))))..... ( -19.80, z-score = -2.42, R) >droSim1.chr3R 10362975 64 + 27517382 -AGUUUACGCUUAUCGGCAUUAGCCGAUGUCAAGGCAUCCGCUGCAU------------------AUCGAUUGGGAAAUUUAC -........((.((((..((((((.(((((....))))).)))).))------------------..)))).))......... ( -17.30, z-score = -1.54, R) >droSec1.super_0 3394254 64 + 21120651 -AGUUUACGCUUAUCGGCAUUAGCCGAUGUCAAGGCAUCCGCUGCAU------------------AUCGAUUGGGAAAUUUAC -........((.((((..((((((.(((((....))))).)))).))------------------..)))).))......... ( -17.30, z-score = -1.54, R) >droEre2.scaffold_4770 273379 64 + 17746568 -AGUUUACGCUUAUCGGCAUUAGCCGAUGUCAAGGCAUCCGCUGCAU------------------AUCGAUUGGGAAAUUUAC -........((.((((..((((((.(((((....))))).)))).))------------------..)))).))......... ( -17.30, z-score = -1.54, R) >droYak2.chr3R 8177361 64 - 28832112 -AGUUUACGCUUAUCGGCAUUAGCCGAUGUCAAGGCAUCCGCUGCAU------------------AUCGAUUGGGAAAUUUAC -........((.((((..((((((.(((((....))))).)))).))------------------..)))).))......... ( -17.30, z-score = -1.54, R) >droAna3.scaffold_13340 13195611 70 - 23697760 --AGUUUAUGCUAUCUGCGUUAGCCGAUGGAGCGGCAUCCGAUGGCCAGGCUC-----------GAUCGAUUUGAAAAUUUAC --.(((.((((.....)))).)))((((.(((((((........)))..))))-----------.)))).............. ( -20.80, z-score = -1.12, R) >dp4.chr2 14624806 81 - 30794189 -ACUUUAUGCUUAUCGGCAUUAGCUGAUGUCCAGGCAUCCGCCAACCGACUAAACUCUUGCAUCGAUCGAUUUGAAAUUUAC- -......(((..((((((....))))))(((..(((....)))....))).........)))((((.....)))).......- ( -15.80, z-score = -1.05, R) >droPer1.super_0 6030063 81 + 11822988 -ACUUUAUGCUUAUCGGCAUUAGCUGAUGUCCAGGCAUCCGCCAACCGACUAAACUCUUGCAUCGAUCGAUUUGAAAUUUAC- -......(((..((((((....))))))(((..(((....)))....))).........)))((((.....)))).......- ( -15.80, z-score = -1.05, R) >droWil1.scaffold_181089 9547397 54 + 12369635 -------AGGUUAUCGGCAUUAGCUGAUGUCAAGGCAUUUGC----C------------------AACGAUUUGAAUUUUUAC -------..(.(((((((....))))))).)..(((....))----)------------------.................. ( -12.70, z-score = -1.85, R) >droVir3.scaffold_12855 9162881 73 + 10161210 -AGUUUAUGUUUAUCGAUACUGGCUGAUGUUGAGGCAUCCGCCA----ACUAAACGUUUGCAU--AUCGAUUUGAAUUUU--- -((((((.....(((((((...((.((((((..(((....))).----....)))))).)).)--)))))).))))))..--- ( -18.00, z-score = -1.99, R) >droMoj3.scaffold_6540 7537580 73 - 34148556 -AGUUUAUGUUUAUCGAUCCUGGCUGAUAUUGAGGCAUCCGCCA----ACUCAACGUUUGCAU--AUCGAUUUGAAUUUU--- -((((((.....((((((....((.((..(((((((....))).----..))))..)).))..--)))))).))))))..--- ( -15.40, z-score = -1.01, R) >droGri2.scaffold_14906 12083687 73 + 14172833 -AGUUUAUGCUUAUCGAUCCUGGCUGAUGUUGAGGCAUCCGCCA----ACUAAACGUUUGCAU--AUCGAUUUGAAUUUU--- -....(((((....((....((((.(((((....))))).))))----......))...))))--)..............--- ( -16.60, z-score = -1.23, R) >consensus _AGUUUACGCUUAUCGGCAUUAGCCGAUGUCAAGGCAUCCGCUGCAU__________________AUCGAUUUGAAAAUUUAC ....................((((.(((((....))))).))))....................................... ( -8.50 = -8.55 + 0.05)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:58:02 2011