| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,064,902 – 4,064,992 |
| Length | 90 |
| Max. P | 0.620095 |

| Location | 4,064,902 – 4,064,992 |
|---|---|
| Length | 90 |
| Sequences | 14 |
| Columns | 90 |
| Reading direction | reverse |
| Mean pairwise identity | 71.12 |
| Shannon entropy | 0.66503 |
| G+C content | 0.54462 |
| Mean single sequence MFE | -27.78 |
| Consensus MFE | -10.52 |
| Energy contribution | -10.36 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.38 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.620095 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 4064902 90 - 27905053 ACCAAUGCUCUCUCUACCUUGCCUCCACUGGAAAUCAACUCCGAUGAGGCCAUGCAGCUGGGCUACAUGCCCAACAGGGACAGCUUUGAG ..(((.(((.(((((.....(((((...((((.......))))..)))))......(.(((((.....))))).)))))).))).))).. ( -31.10, z-score = -2.75, R) >droWil1.scaffold_181130 1222477 90 + 16660200 UCGAAUGCCCUGGCCAGAUUACCUCCACUGGAGAUAAGCAGUGACGAGGCCAUGCAGUUAGGUUAUAUGCCAAGUAGAGACAGUUUUGAA ((((((((..(((((.((.....))(((((........)))))....))))).)))....(((.....))).............))))). ( -21.20, z-score = -0.40, R) >droMoj3.scaffold_6540 24889809 90 + 34148556 GCCAAUGCGCUUGCCCGCCUGCCACCCCUAGAUAUAACCAAUGAAGAAGCGCUGCAACUGGGCUACAUGCCCAAUCGUGAUAGCUUUGAG ((....(((......)))..)).......................(((((....((..(((((.....)))))....))...)))))... ( -17.30, z-score = 0.83, R) >droVir3.scaffold_13047 10094056 90 - 19223366 GCCAAUGCACUUGCCCGCCUGCCACCGCUAGAGAUAAGCAACGAGGAAGCGCUUCAACUGGGCUAUAUGCCCAAUCGUGAUAGCUUUGAA ((....))..((((...(((((....)).)).)....))))....(((((...(((..(((((.....)))))....)))..)))))... ( -20.60, z-score = 0.87, R) >droGri2.scaffold_14906 6805230 90 + 14172833 GCCAAUGCGCUUGCCCGCCUGCCACCCCUCGAGAUAAGUAGUGAGGAGGCGCUGCAAUUGGGCUACAUGCCCAAUCGGGAUAGUUUUGAG ..(((.((.....(((((((.((((..((.......))..))).).))))......(((((((.....))))))).)))...)).))).. ( -30.50, z-score = -0.80, R) >droPer1.super_0 7453290 90 - 11822988 GCCAGUGCCAUGGCCAAUCUGCCACCUCUGGAGAUCAACGCCGACGAGGCGGCGCAGCUGGGCUACAUGCCCAACAGGGACAGCUUUGAG ((..(((((.((((......))))(((((((.(.....).)))..)))).))))).))(((((.....))))).(((((....))))).. ( -32.20, z-score = -0.24, R) >dp4.chr2 17791231 90 + 30794189 GCCAGUGCCAUGGCCAAUCUGCCACCUCUGGAGAUCAACGCCGACGAGGCGGCGCAGCUGGGCUACAUGCCCAACAGGGACAGCUUUGAG ((..(((((.((((......))))(((((((.(.....).)))..)))).))))).))(((((.....))))).(((((....))))).. ( -32.20, z-score = -0.24, R) >droAna3.scaffold_13340 21523166 90 - 23697760 ACUAGUGCUCUGGCCAGUCUCCCGCCACUAGACAUCAACACUGACGAAGCUAUGCAGCUGGGCUACAUGCCCAACCGAGAUAGCUUCGAA .((((((...(((........))).)))))).............(((((((((.(.(.(((((.....))))).).)..))))))))).. ( -30.50, z-score = -2.70, R) >droYak2.chr3R 8140614 90 - 28832112 ACCAAUGCACUCUCCACUUUGCCUCCAUUGGAAAUCAACUCCGAUGAGGCCAUGCAGCUGGGCUACAUGCCCAACAGGGACAGCUUCGAA ......((..((.((.....(((((.((((((.......)))))))))))......(.(((((.....))))).).))))..))...... ( -28.50, z-score = -2.03, R) >droEre2.scaffold_4770 237080 90 + 17746568 ACCAAUGCAUUGUCCACCUUGCCUCCCUUGGAUAUCAACUCCGAUGAGGCCAUGCAGCUGGGCUACAUGCCCAACAGGGACAGCUUUGAA ..(((.((..(((((.....(((((..(((((.......))))).)))))......(.(((((.....))))).)..))))))).))).. ( -31.60, z-score = -2.35, R) >droSec1.super_0 3356596 90 + 21120651 ACUAAUGCUCUCUCUACCUUGCCUCCAUUGGAAAUCAACUCCGAUGAGGCGAUGCAGCUGGGCUACAUGCCCAACAGGGACAGCUUUGAG ......(((.(((((...(((((((.((((((.......)))))))))))))....(.(((((.....))))).)))))).)))...... ( -34.80, z-score = -4.08, R) >droSim1.chr3R 10323889 90 + 27517382 ACCAAUGCUCUCUCUACCUUGCCUCCAUUGGAAAUCAACUCCGAUGAGGCGAUGCAGCUGGGCUACAUGCCCAACAGGGACAGCUUUGAG ..(((.(((.(((((...(((((((.((((((.......)))))))))))))....(.(((((.....))))).)))))).))).))).. ( -36.90, z-score = -4.53, R) >anoGam1.chr2R 30223349 88 + 62725911 GCUGCUGAUACAG--AAGCUGCCGCCGAUGGACUGUACCGCAGAGGAGGCCGCCGCUCUCGGAUACAUGCCGAAUCGGGACGACUUUGAG ((.(((.......--.))).))..(((((((..(((((((.((((..((...)).))))))).))))..))..)))))............ ( -26.10, z-score = 0.19, R) >triCas2.ChLG7 5253823 90 - 17478683 CCUGCAGUGAUUGCAAGGUUGCCACCUUUAGAUAUAACUCUAGAAGAAGCUCAAUUGUUAGGGUAUAAACCACAUAGGGACGAUUUUGAA (((((((...))))..((((...(((((((((......))))))...(((......)))..)))...))))....)))............ ( -15.40, z-score = 0.90, R) >consensus ACCAAUGCUCUGGCCAGCCUGCCACCACUGGAGAUCAACACCGAUGAGGCGAUGCAGCUGGGCUACAUGCCCAACAGGGACAGCUUUGAG .............................................(((((........(((((.....))))).........)))))... (-10.52 = -10.36 + -0.16)
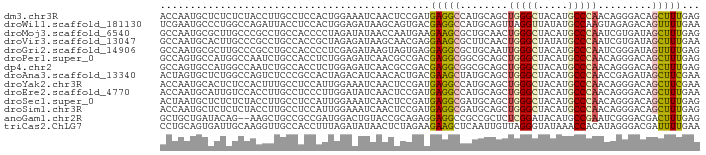


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:57:59 2011