| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,029,660 – 4,029,763 |
| Length | 103 |
| Max. P | 0.537262 |

| Location | 4,029,660 – 4,029,763 |
|---|---|
| Length | 103 |
| Sequences | 8 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 75.57 |
| Shannon entropy | 0.46861 |
| G+C content | 0.43460 |
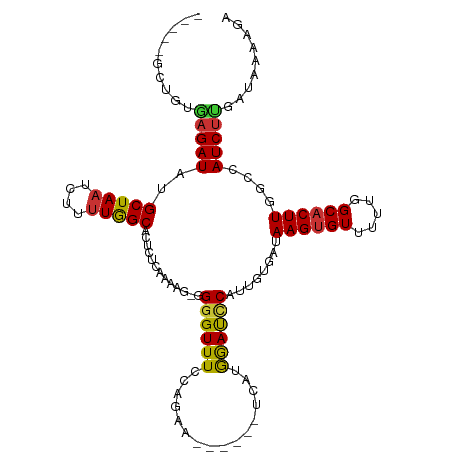
| Mean single sequence MFE | -31.83 |
| Consensus MFE | -15.35 |
| Energy contribution | -14.80 |
| Covariance contribution | -0.54 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.537262 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
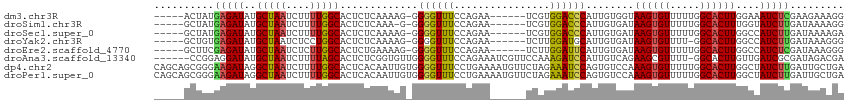
>dm3.chr3R 4029660 103 + 27905053 -----ACUAUGAGAUAUGCUAAUCUUUUGGCACUCUCAAAAG-GGGGUUUCCAGAA------UCGUGGACCCAUUGUGGUAAGUGUUUUUGGCACUUGGAAAUCUCGAAGAAAGG -----....(((((..((((((....)))))).)))))....-(((((((((....------......(((......)))((((((.....)))))))))))))))......... ( -30.10, z-score = -1.66, R) >droSim1.chr3R 10289486 102 - 27517382 -----GCUAUGAGAUAUGCUAAUCUUUUGGCACUCUCAAA-G-GGGGUUUCCAGAA------UCGUGGACCCAUUGUGAUAAGUGUUUUUGGCACUUUGGUAUCUUGAUAAAAGG -----.((.(((((..((((((....)))))).))))).)-)-.(((((..(....------..)..)))))((((.(((((((((.....)))))....)))).))))...... ( -26.30, z-score = -0.83, R) >droSec1.super_0 3321490 103 - 21120651 -----GCUAUGAGAUAUGCUAAUCUUUUGGCACUCUCAAAAG-GGGGUUUCCAGAA------UCGUGGACCCAUUGUGAUAAGUGUUUUUGGCACUUGGCCAUCUUGAUAAAAGA -----....(((((..((((((....)))))).)))))...(-((((((..(....------..)..))))).......(((((((.....))))))).)).((((.....)))) ( -28.90, z-score = -1.23, R) >droYak2.chr3R 8103899 102 + 28832112 -----GCUGUGAGAUAUGCUAAUCUCCUGGCACUCUCAAAAG-GGGGUUUCCAGAA------UCUUGGAUGCAUUGUGAUAAGUGUUUU-GGCACUUGGCCAUCUUGAUAAAGGG -----((((.(((((......))))).))))((((((....)-))))).(((((..------..)))))..((..(((.(((((((...-.)))))))..)))..))........ ( -27.60, z-score = -0.21, R) >droEre2.scaffold_4770 198051 103 - 17746568 -----GCUUCGAGAUAUGCUAAUCUCUUGGCACUCUGAAAAG-GGGGUUUCCAGAA------UCUUGGAUUCAUUGUGAUAAGUGUUUUUGGCACUUGGCCAUCUCGAUAAAGGG -----...(((((((.((((((....))))))((((....))-))(((.(((((..------..)))))..........(((((((.....)))))))))))))))))....... ( -30.80, z-score = -1.72, R) >droAna3.scaffold_13340 21484368 108 + 23697760 ------CCGGAGGAUAUGCUAAUCUUUUAGCACUCUCGGUGUUGGGGUUUCCAGAAAUCGUUCCAAAGAUCCAUUGUCAGAAGCGUUUU-GGCACUUGUUGAUCGCGAUAGACGA ------((((((....((((((....))))))))).))).....((((((...(((....)))...)))))).(((((....(((..(.-.((....))..).)))....))))) ( -27.90, z-score = 0.03, R) >dp4.chr2 17755431 115 - 30794189 CAGCAGCGGGAAGAUAGGCUAAUCUUUUGGCACUCACAAUUGUGGGGUUUCCUGAAAAUGUUCUAGAAAUCCAGUGUCCAAAGUGUUUUUGGCACUUGGCUAUCUUGAUUGCUGA ...((((((.(((((((.((((...((((((((((((....)))(((((((..(((....)))..))))))))))).)))))((((.....)))))))))))))))..)))))). ( -41.50, z-score = -3.37, R) >droPer1.super_0 7416280 115 + 11822988 CAGCAGCGGGAAGAUAGGCUAAUCUUUUGGCACUCACAAUUGUGGGGUUUCCUGAAAAUGUUCUAGAAAUCCAGUGUCCAAAGUGUUUUUGGCACUUGGCUAUCUUGAUUGCUGA ...((((((.(((((((.((((...((((((((((((....)))(((((((..(((....)))..))))))))))).)))))((((.....)))))))))))))))..)))))). ( -41.50, z-score = -3.37, R) >consensus _____GCUGUGAGAUAUGCUAAUCUUUUGGCACUCUCAAAAG_GGGGUUUCCAGAA______UCAUGGAUCCAUUGUGAUAAGUGUUUUUGGCACUUGGCCAUCUUGAUAAAAGA ..........(((((..(((((....))))).............((((((................))))))........((((((.....))))))....)))))......... (-15.35 = -14.80 + -0.54)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:57:53 2011