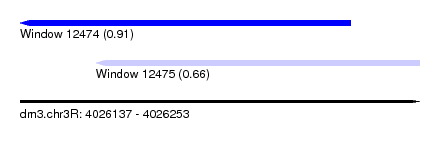
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,026,137 – 4,026,253 |
| Length | 116 |
| Max. P | 0.914496 |

| Location | 4,026,137 – 4,026,233 |
|---|---|
| Length | 96 |
| Sequences | 9 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 75.11 |
| Shannon entropy | 0.51203 |
| G+C content | 0.48954 |
| Mean single sequence MFE | -23.59 |
| Consensus MFE | -15.79 |
| Energy contribution | -15.61 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.24 |
| SVM RNA-class probability | 0.914496 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
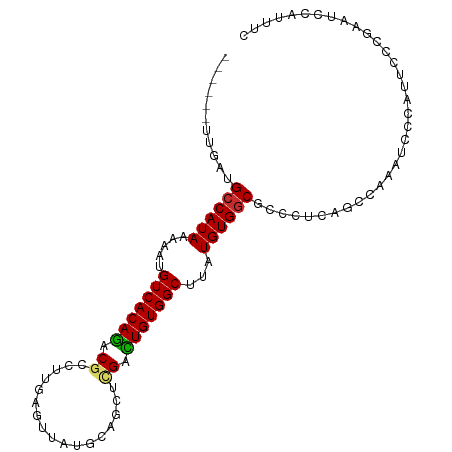
>dm3.chr3R 4026137 96 - 27905053 ------UUGAUGCCAUAAAAAUGUCACAGACGCCUUGAGUUAUGCAGCUCGACUGUGGCUUAUGUGGCGCCUACAGCCAAAUCCCAUUCCCGAAUCCAUUUC ------..(((((((((.....(((((((.....(((((((....))))))))))))))...))))))((.....))...)))................... ( -22.70, z-score = -1.20, R) >droSim1.chr3R 10285976 96 + 27517382 ------UUGAUGCCAUAAAAAUGUCACAGACGCCUUGAGUUAUGCAGCUCGACUGUGGCUUAUGUGGCGCUCUCAGCCAAAUCCCAUUCCCGAAUCCAUUUC ------..(((((((((.....(((((((.....(((((((....))))))))))))))...))))))).)).............................. ( -23.50, z-score = -1.29, R) >droSec1.super_0 3317997 96 + 21120651 ------UUGAUGCCAUAAAAAUGUCACAGACGCCUUGAGUUAUGCAGCUCGACUGUGGCUUAUGUGGCGCCCUCAGCCAAAUCCCAUUCCCGAAUCCAUUUC ------(((((((((((.....(((((((.....(((((((....))))))))))))))...)))))))...)))).......................... ( -23.20, z-score = -1.38, R) >droYak2.chr3R 8100341 96 - 28832112 ------UUGAUGCCAUAAAAAUGUCACAGACGCCUUGAGUUAUGCAGCUCGACUGUGGCUUAUGUGGCGCCCUCAGCCAAAUCCCAUUCCCGAAUCCAUUUC ------(((((((((((.....(((((((.....(((((((....))))))))))))))...)))))))...)))).......................... ( -23.20, z-score = -1.38, R) >droEre2.scaffold_4770 194564 96 + 17746568 ------UUGAUGCCAUAAAAAUGUCACAGACGCCUUGAGUUAUGCAGCUCGACUGUGGCUUAUGUGGCGCCCUCAGCCAAAUCCCAUUCCCGGAUCCAUUUC ------(((((((((((.....(((((((.....(((((((....))))))))))))))...)))))))...))))....((((.......))))....... ( -24.90, z-score = -1.19, R) >droAna3.scaffold_13340 21480239 96 - 23697760 ------UUGAUGCCAUAAAAAUGUCACAGACGCCUUGAGUUAUGCAGCCUGACUGUGGCUUAUGUGGCGCCCGCCGUAAAAUCCCAUUCCGAAAUCCAUUCC ------.....((((((.....(((((((.((..(((.......)))..)).)))))))...)))))).................................. ( -18.60, z-score = 0.02, R) >droWil1.scaffold_181130 14142822 99 - 16660200 UACAUUUUGCUGCCAUAAAAAUGUCACAGACGCCUUGAAUUAUGG-GCGUGGUUGUGGCUUGUGUGUCUCUGUGUGUGUGUGUGUGUUCCCAAUCCUACU-- (((((..(((.(.((((.....(((((((((((((........))-))))..)))))))...)))).)...)))...)))))..................-- ( -23.10, z-score = -0.05, R) >droGri2.scaffold_14906 6769341 81 + 14172833 -------CUAUGCCAUAAAAAUGUCACAAGCUCCUUGAGUUAUGU-GCGUGGUUGUGGCUUGUGUGGCGC-CUCGACACCCCCUCGUUUC------------ -------....(((((((..(((.(((((((((...))))).)))-))))..)))))))..((((.(...-..).))))...........------------ ( -23.80, z-score = -2.02, R) >droVir3.scaffold_13047 10053209 87 - 19223366 -UGCAUACUAUGCCAUAAAAAUGUCACAAACUCCUUGAGUUAUGU-GCGUGGUUGUGGCUUGUGUGGCGC-UGAGGUAGCCCCACCCCUC------------ -.....((...(((((((..(((.(((((((((...))))).)))-))))..)))))))..))((((.((-((...)))).)))).....------------ ( -29.30, z-score = -2.37, R) >consensus ______UUGAUGCCAUAAAAAUGUCACAGACGCCUUGAGUUAUGCAGCUCGACUGUGGCUUAUGUGGCGCCCUCAGCCAAAUCCCAUUCCCGAAUCCAUUUC ...........((((((.....(((((((.((.................)).)))))))...)))))).................................. (-15.79 = -15.61 + -0.18)


| Location | 4,026,159 – 4,026,253 |
|---|---|
| Length | 94 |
| Sequences | 7 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 85.51 |
| Shannon entropy | 0.26711 |
| G+C content | 0.44157 |
| Mean single sequence MFE | -27.44 |
| Consensus MFE | -20.59 |
| Energy contribution | -20.43 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.658703 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 4026159 94 - 27905053 GCGGUGAAUAAAAUAUAUA---------------UUUGAUGCCAUAAAAAUGUCACAGACGCCUUGAGUUAUGCAGCUCGACUGUGGCUUAUGUGGCGCCUACAGCCAA ..((((((((.......))---------------)))...((((((.....(((((((.....(((((((....))))))))))))))...)))))).......))).. ( -25.70, z-score = -1.18, R) >droSim1.chr3R 10285998 94 + 27517382 GCGGCGAAUAAAAUAUAUA---------------UUUGAUGCCAUAAAAAUGUCACAGACGCCUUGAGUUAUGCAGCUCGACUGUGGCUUAUGUGGCGCUCUCAGCCAA ..((((((((.......))---------------)))(((((((((.....(((((((.....(((((((....))))))))))))))...))))))).))...))).. ( -29.00, z-score = -1.98, R) >droSec1.super_0 3318019 94 + 21120651 GCGGCGAAUAAAAUAUAUA---------------UUUGAUGCCAUAAAAAUGUCACAGACGCCUUGAGUUAUGCAGCUCGACUGUGGCUUAUGUGGCGCCCUCAGCCAA ..((((((((.......))---------------)))...((((((.....(((((((.....(((((((....))))))))))))))...)))))).......))).. ( -28.10, z-score = -1.74, R) >droYak2.chr3R 8100363 94 - 28832112 CCGGCGAAUAAAAUAUAUA---------------UUUGAUGCCAUAAAAAUGUCACAGACGCCUUGAGUUAUGCAGCUCGACUGUGGCUUAUGUGGCGCCCUCAGCCAA ..((((((((.......))---------------)))...((((((.....(((((((.....(((((((....))))))))))))))...)))))).......))).. ( -28.10, z-score = -2.17, R) >droEre2.scaffold_4770 194586 94 + 17746568 CCGGCGAAUAAAAUAUAUA---------------UUUGAUGCCAUAAAAAUGUCACAGACGCCUUGAGUUAUGCAGCUCGACUGUGGCUUAUGUGGCGCCCUCAGCCAA ..((((((((.......))---------------)))...((((((.....(((((((.....(((((((....))))))))))))))...)))))).......))).. ( -28.10, z-score = -2.17, R) >droAna3.scaffold_13340 21480261 94 - 23697760 CCAGCGAAUAAAAUAUAUA---------------UUUGAUGCCAUAAAAAUGUCACAGACGCCUUGAGUUAUGCAGCCUGACUGUGGCUUAUGUGGCGCCCGCCGUAAA ...(((....((((....)---------------)))...((((((.....(((((((.((..(((.......)))..)).)))))))...))))))...)))...... ( -20.50, z-score = -0.11, R) >droWil1.scaffold_181130 14142842 108 - 16660200 GUAACGUAUAAAAUAUAUGUACAUAUACAUACAUUUUGCUGCCAUAAAAAUGUCACAGACGCCUUGAAUUAUG-GGCGUGGUUGUGGCUUGUGUGUCUCUGUGUGUGUG (((.((((((...)))))))))...(((((((((...(..(((((((....(((((((((((((........)-)))))..)))))))))))).))..).))))))))) ( -32.60, z-score = -2.09, R) >consensus GCGGCGAAUAAAAUAUAUA_______________UUUGAUGCCAUAAAAAUGUCACAGACGCCUUGAGUUAUGCAGCUCGACUGUGGCUUAUGUGGCGCCCUCAGCCAA ..(((..(((.....)))......................((((((.....(((((((.....(((((((....))))))))))))))...)))))).......))).. (-20.59 = -20.43 + -0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:57:53 2011