
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,025,665 – 4,025,776 |
| Length | 111 |
| Max. P | 0.842500 |

| Location | 4,025,665 – 4,025,776 |
|---|---|
| Length | 111 |
| Sequences | 9 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 64.59 |
| Shannon entropy | 0.73610 |
| G+C content | 0.54696 |
| Mean single sequence MFE | -33.41 |
| Consensus MFE | -15.85 |
| Energy contribution | -14.44 |
| Covariance contribution | -1.41 |
| Combinations/Pair | 1.83 |
| Mean z-score | -0.79 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.842500 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
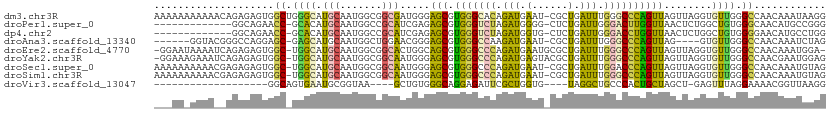
>dm3.chr3R 4025665 111 + 27905053 AAAAAAAAAAACAGAGAGUGGCUGGGCAUGCAAUGGCGGCGAUGGGAGCGUGGGCACAGAUGAAU-CGCUGAUUUGGGCCCAGUUAGUUAGGUGUUGGGCCAACAAAUAAGG ...............((.(((((((((...(((...(((((((..(.((....)).)......))-)))))..))).))))))))).)).(((.....)))........... ( -32.00, z-score = -1.33, R) >droPer1.super_0 7411912 97 + 11822988 -------------GGCAGAACC-GCACAUGCAAUGGCCGCAUCGAGAGCGUGGGUCUAGAUGGGG-CUCUGAUUGGGACUUGGUUAACUCUGGCUGUGGGCAACAUGCCGGG -------------((((.....-((....))..((.(((((.(.((((...((((((.....)))-)))((((..(...)..)))).)))).).))))).))...))))... ( -31.70, z-score = 0.39, R) >dp4.chr2 17751038 97 - 30794189 -------------GGCAGAACC-GCACAUGCAAUGGCCGCAUCGAGAGCGUGGGUCUAGAUGGUG-CUCUGAUUGGGACCUGGUUAACUCUGGCUGUGGGGAACAUGCCUGG -------------((((...((-.((((.((...((((.((((....).))))))).....(((.-(((.....))))))............)))))).))....))))... ( -31.00, z-score = 0.60, R) >droAna3.scaffold_13340 21479420 100 + 23697760 ------GGUACGGGCCAGGAGC-GAGCAUGCAAUGGCUGGAACGGGAGCGUGGGCCAAGAUGAAU-CGCUGAUUUGGGCCCAGUUAG----GUGUUGGGCCAACAAAUCUAG ------(((....)))...(((-((.(((....(((((...(((....))).)))))..)))..)-))))(((((((((((((....----...)))))))..))))))... ( -35.80, z-score = -1.16, R) >droEre2.scaffold_4770 194038 109 - 17746568 -GGAAUAAAAUCAGAGAGUGGC-UGGCAUGCAAUGGCGGCACUGGCAGCGUGGGCCCAGAUGAAUGCGCUGAUUUGGGCCCAGUUAGUUAGGUGUUGGGCCAACAAAUGGA- -.........(((.....((((-..((((((....))....((((((((.(((((((((((.(......).)))))))))))))).)))))))))..).))).....))).- ( -40.80, z-score = -1.59, R) >droYak2.chr3R 8099859 110 + 28832112 -GGAAAGAAAUCAGAGAGUGGC-UGGCAUGCAAUGGCGGCAAUGGGAGCGUGGGCCCAGAUGAGUACGCUGAUUUGGGCCCAGUUAGUUAGGUGUUGGGCCAACGAAUGGAG -.........(((.....((((-..((((....((((.((.......)).(((((((((((.((....)).))))))))))))))).....))))..).))).....))).. ( -39.20, z-score = -2.41, R) >droSec1.super_0 3317553 110 - 21120651 AAAAAAAAAACGAGAGAGUGGC-UGGCAUGCAAUGGCGGCAAUGGGAGCGUGGGCCCAGAUGAAU-CGCUGAUUUGGACCCAGUUAGUUAGGUGUUGGGCCAACAAAUGUAG .........(((.....((.((-((.(((...))).)))).)).....))).(((((((....((-((((((((.(....))))))))..))).)))))))........... ( -29.70, z-score = -0.17, R) >droSim1.chr3R 10285509 110 - 27517382 AAAAAAAAAACGAGAGAGUGGC-UGGCAUGCAAUGGCGGCAAUGGGAGCGUGGGCCCAGAUGAAU-CGCUGAUUUGGGCCCAGUUAGUUAGGUGUUGGGCCAACAAAUGUAG ..........(....)..((((-..((((....((((.((.......)).(((((((((((.(..-...).))))))))))))))).....))))..).))).......... ( -33.60, z-score = -0.97, R) >droVir3.scaffold_13047 10052270 84 + 19223366 -------------------GGCAGUGAAUGCGGUAA----GCUGUGGGCAGGAGAUUCGCUGGUG----UAGGCUGCCCACUGCUAGCU-GAGUUUAGGAAAACGGUUAAGG -------------------.....(((((.((((.(----((.((((((((...((........)----)...)))))))).))).)))-).)))))............... ( -26.90, z-score = -0.49, R) >consensus ___AA_AAAA__AGAGAGUGGC_GGGCAUGCAAUGGCGGCAAUGGGAGCGUGGGCCCAGAUGAAU_CGCUGAUUUGGGCCCAGUUAGUUAGGUGUUGGGCCAACAAAUGUAG ....................((.((((.(((.......))).....(((.(((((((.(((.(......).))).))))))))))........)))).))............ (-15.85 = -14.44 + -1.41)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:57:51 2011