| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,025,502 – 4,025,592 |
| Length | 90 |
| Max. P | 0.635692 |

| Location | 4,025,502 – 4,025,592 |
|---|---|
| Length | 90 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.49 |
| Shannon entropy | 0.49416 |
| G+C content | 0.51705 |
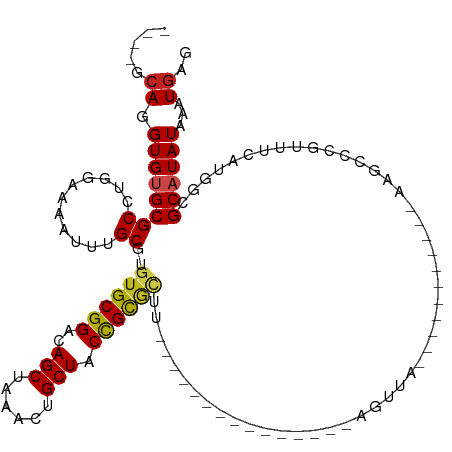
| Mean single sequence MFE | -32.00 |
| Consensus MFE | -16.75 |
| Energy contribution | -16.53 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.635692 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
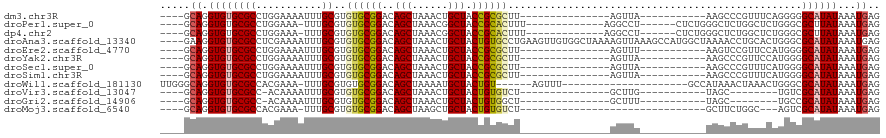
>dm3.chr3R 4025502 90 + 27905053 ----GCAGGUGUGCGCCUGGAAAAUUUGCGUGUGCGGACAGCUAAACUGCUACCGCGCUU---------------AGUUA-----------AAGCCCGUUUCAGGGGGCAUAUAAAUGAG ----.((.((((((.(((.((((.(((((..((((((..(((......))).))))))..---------------.).))-----------)).....)))).))).))))))...)).. ( -30.90, z-score = -0.93, R) >droPer1.super_0 7411755 96 + 11822988 ----GCAGGUGUGCGCCUGGAAA-UUUGCGUGUGCGGACAGCUAAACGGCUACCGCACUUU-------------AGGCCU------CUCUGGGCUCUGGCUCUGGGCGCUUAUAAAUGAG ----.((.(((.(((((..((..-...((..((((((..((((....)))).))))))..(-------------((((((------....)))).)))))))..))))).)))...)).. ( -38.10, z-score = -1.71, R) >dp4.chr2 17750889 96 - 30794189 ----GCAGGUGUGCGCCUGGAAA-UUUGCGUGUGCGGACAGCUAAACGGCUACCGCACUUU-------------AGGCCU------CUCUGGGCUCUGGCUCUGGGCGCUUAUAAAUGAG ----.((.(((.(((((..((..-...((..((((((..((((....)))).))))))..(-------------((((((------....)))).)))))))..))))).)))...)).. ( -38.10, z-score = -1.71, R) >droAna3.scaffold_13340 21479239 116 + 23697760 ----GAAGGUGUGCGCCUCGAAAAUUUGCGUGUGCGGACAGCUAAACUGCUACUGUGCCUGAAGUUGUGGCUAAAAGUUAAAGCCAUGGCUAAAACCUGCACUGGGCGCAUAUAAAUGAG ----....((((((((((((........)).((((((..(((......)))...........(((..(((((.........)))))..))).....)))))).))))))))))....... ( -39.50, z-score = -2.41, R) >droEre2.scaffold_4770 193883 90 - 17746568 ----GCAGGUGUGCGCCUGGAAAAUUUGCGUGUGCGGACAGCUAAACUGCUACCGCGCUU---------------AGUUU-----------AAGUCCGUUCCAUGGGGCAUAUAAAUGAG ----.((.((((((.(((((((.((((((..((((((..(((......))).))))))..---------------.))..-----------))))...))))).)).))))))...)).. ( -33.00, z-score = -2.10, R) >droYak2.chr3R 8099700 90 + 28832112 ----GCAGGUGUGCGCCUGGAAAAUUUGCGUGUGCGGACAGCUAAACUGCUACCGCGCUU---------------AGUUA-----------AAGCCCGUUCCAUGGGGCAUAUAAAUGAG ----.((.((((((.(((((((..(((((..((((((..(((......))).))))))..---------------.).))-----------)).....))))).)).))))))...)).. ( -33.00, z-score = -1.78, R) >droSec1.super_0 3317396 90 - 21120651 ----GCAGGUGUGCGCCUGGAAAAUUUGCGUGUGCGGACAGCUAAACUGCUACCGCGCUU---------------AGUUA-----------AAGCCCGUUUCAUGGGGCAUAUAAAUGAG ----.((.((((((.(((((((..(((((..((((((..(((......))).))))))..---------------.).))-----------)).....))))).)).))))))...)).. ( -30.10, z-score = -0.95, R) >droSim1.chr3R 10285354 90 - 27517382 ----GCAGGUGUGCGCCUGGAAAAUUUGCGUGUGCGGACAGCUAAACUGCUACCGCGCUU---------------AGUUA-----------AAGCCCGUUUCAUGGGGCAUAUAAAUGAG ----.((.((((((.(((((((..(((((..((((((..(((......))).))))))..---------------.).))-----------)).....))))).)).))))))...)).. ( -30.10, z-score = -0.95, R) >droWil1.scaffold_181130 14136103 92 + 16660200 UUGGGCAGGUGUGCGCCACGAAA-UUUGCGUGUGCGGACAGCUAAAAUGCUACUGU------AGUUU---------------------GCCAUAAACUAAACUGGGCGCAUAUAAAUGAG ....((((.(((.(((((((...-....)))).))).))).)).....(((....(------(((((---------------------(...))))))).....)))))........... ( -26.10, z-score = -0.40, R) >droVir3.scaffold_13047 10052129 81 + 19223366 ----GCAGGUGUGCGCC-ACAAAAUUUGCGUGUGCGGACAGCUAAACUGCUACUGUGUCU---------------GCUUG-----------UAGC--------UGUCGCAUAUAAAUGAG ----.((.(((((((((-((.........))).)).((((((((....((..........---------------))...-----------))))--------))))))))))...)).. ( -27.10, z-score = -1.06, R) >droGri2.scaffold_14906 6768529 81 - 14172833 ----GCAGGUGUGCGCC-ACAAAAUUUGCGUGUGCGGACAGCUAAACUGCUACUGUGGCU---------------GCUUU-----------UAGC--------UGCCGCAUAUAAAUGAG ----((((.((((...)-)))....))))((((((((.((((((((..(((.....))).---------------...))-----------))))--------))))))))))....... ( -30.20, z-score = -1.74, R) >droMoj3.scaffold_6540 24843897 81 - 34148556 ----GCAGGUGUGCGCCACGAAA-UUUGCGUGUGCGGACAGCUAAGCUGCUACUGUGUCU-------------------------------GCUUCUGGC---AGUCGCAUAUAAAUGAG ----((((.(((.(((((((...-....)))).))).))).))..))......((((.((-------------------------------((.....))---)).)))).......... ( -27.80, z-score = -0.48, R) >consensus ____GCAGGUGUGCGCCUGGAAAAUUUGCGUGUGCGGACAGCUAAACUGCUACCGCGCUU_______________AGUUA___________AAGCCCGUUUCAUGGCGCAUAUAAAUGAG .....((.((((((((...........))..((((((..(((......))).)))))).................................................))))))...)).. (-16.75 = -16.53 + -0.22)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:57:50 2011