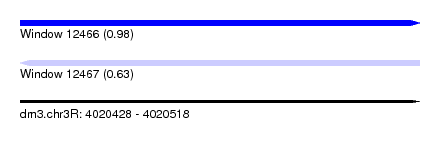
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 4,020,428 – 4,020,518 |
| Length | 90 |
| Max. P | 0.979783 |

| Location | 4,020,428 – 4,020,518 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 68.11 |
| Shannon entropy | 0.48411 |
| G+C content | 0.49443 |
| Mean single sequence MFE | -18.83 |
| Consensus MFE | -13.50 |
| Energy contribution | -13.10 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.979783 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 4020428 90 + 27905053 GCAUUCGGGGCAAUACAACAAAAACAAUAAAACGUGGCAGGCGCAUCCUGCCCCAUUU------------------------UCGGCCCCCACUUCCAUUUUAUCCGUGCAACC ((((..(((((............((........))((((((.....))))))......------------------------...)))))................)))).... ( -20.67, z-score = -0.60, R) >droAna3.scaffold_13340 21474832 77 + 23697760 --GGCCGAGGCAAUACAACAAAAACAAUAAAACGUGGCAGGCGCAUCCUGCC---CAU--------CCCGCCUC-----C--UUUAUCCUUCCAACC----------------- --....(((((............((........))((((((.....))))))---...--------...)))))-----.--...............----------------- ( -18.20, z-score = -2.01, R) >droEre2.scaffold_4770 188795 73 - 17746568 GCAUUCAGGGCAAUACAACAAAAACAAUAAAACGUGGCAGGCGCAUCCUGCCACCCAU------------------------UUUAUCCUUGCAACC----------------- .....(((((................((((((.((((((((.....))))))))...)------------------------)))))))))).....----------------- ( -17.79, z-score = -2.11, R) >droYak2.chr3R 8094484 97 + 28832112 GCAUUCAGGGCAAUACAACAAAAACAAUAAAACGUGGCAGGCGCAUCCUGCCACCCAUUUUCUCACCCCACUCCUCCCUCAUUUUAUCCUUGCAACC----------------- (((...((((.....).................((((((((.....)))))))).................................))))))....----------------- ( -16.90, z-score = -1.80, R) >droSec1.super_0 3312386 92 - 21120651 GCAUUCGAGGCAAUACAACAAAAACAAUAAAACGUGGCAGGCGCAUCCUGCCACCCAUUUUUGGCCCCCACUUC-----CAUUUUAUCCUUGCAACC----------------- .....(((((.......................((((((((.....))))))))(((....)))..........-----........))))).....----------------- ( -20.60, z-score = -1.86, R) >consensus GCAUUCGGGGCAAUACAACAAAAACAAUAAAACGUGGCAGGCGCAUCCUGCCACCCAU________CCC_C__C_____C__UUUAUCCUUGCAACC_________________ .....(((((.............((........))((((((.....))))))...................................)))))...................... (-13.50 = -13.10 + -0.40)

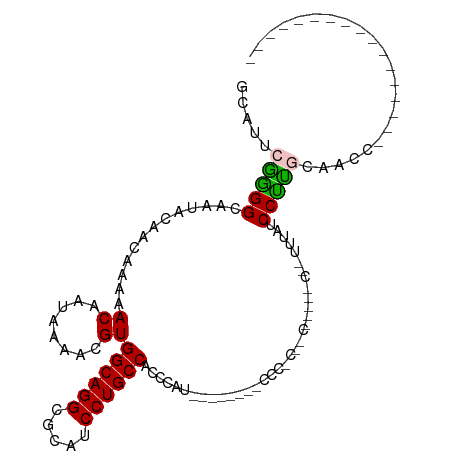

| Location | 4,020,428 – 4,020,518 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 68.11 |
| Shannon entropy | 0.48411 |
| G+C content | 0.49443 |
| Mean single sequence MFE | -26.38 |
| Consensus MFE | -15.24 |
| Energy contribution | -15.00 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.16 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.632862 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 4020428 90 - 27905053 GGUUGCACGGAUAAAAUGGAAGUGGGGGCCGA------------------------AAAUGGGGCAGGAUGCGCCUGCCACGUUUUAUUGUUUUUGUUGUAUUGCCCCGAAUGC ....((((((....((((..((..(((((..(------------------------(((((.((((((.....)))))).))))))...)))))..)).))))...)))..))) ( -26.50, z-score = 0.02, R) >droAna3.scaffold_13340 21474832 77 - 23697760 -----------------GGUUGGAAGGAUAAA--G-----GAGGCGGG--------AUG---GGCAGGAUGCGCCUGCCACGUUUUAUUGUUUUUGUUGUAUUGCCUCGGCC-- -----------------(((((..(((((((.--.-----((((((((--------(((---((((((.....)))))).))))...)))))))..))))....))))))))-- ( -24.60, z-score = -0.96, R) >droEre2.scaffold_4770 188795 73 + 17746568 -----------------GGUUGCAAGGAUAAA------------------------AUGGGUGGCAGGAUGCGCCUGCCACGUUUUAUUGUUUUUGUUGUAUUGCCCUGAAUGC -----------------(((.((((((((((.------------------------..(.((((((((.....)))))))).)....))))))))))......)))........ ( -24.90, z-score = -2.24, R) >droYak2.chr3R 8094484 97 - 28832112 -----------------GGUUGCAAGGAUAAAAUGAGGGAGGAGUGGGGUGAGAAAAUGGGUGGCAGGAUGCGCCUGCCACGUUUUAUUGUUUUUGUUGUAUUGCCCUGAAUGC -----------------................(.((((...((((.((..(((((((((((((((((.....))))))))...))))).))))..)).)))).)))).).... ( -27.70, z-score = -1.51, R) >droSec1.super_0 3312386 92 + 21120651 -----------------GGUUGCAAGGAUAAAAUG-----GAAGUGGGGGCCAAAAAUGGGUGGCAGGAUGCGCCUGCCACGUUUUAUUGUUUUUGUUGUAUUGCCUCGAAUGC -----------------((((((((.((...((((-----(((.......(((....)))((((((((.....)))))))).)))))))....)).)))))..)))........ ( -28.20, z-score = -1.10, R) >consensus _________________GGUUGCAAGGAUAAA__G_____G__G_GGG________AUGGGUGGCAGGAUGCGCCUGCCACGUUUUAUUGUUUUUGUUGUAUUGCCCCGAAUGC .................(((....(.((((((..............................((((((.....))))))(((......))).)))))).)...)))........ (-15.24 = -15.00 + -0.24)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:57:46 2011