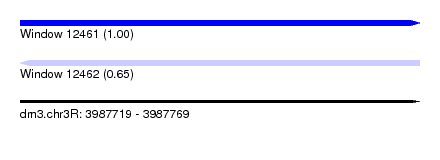
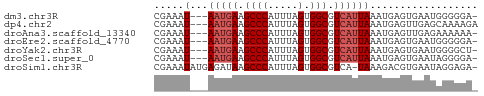
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 3,987,719 – 3,987,769 |
| Length | 50 |
| Max. P | 0.996333 |

| Location | 3,987,719 – 3,987,769 |
|---|---|
| Length | 50 |
| Sequences | 7 |
| Columns | 54 |
| Reading direction | forward |
| Mean pairwise identity | 84.36 |
| Shannon entropy | 0.29233 |
| G+C content | 0.39663 |
| Mean single sequence MFE | -9.27 |
| Consensus MFE | -6.36 |
| Energy contribution | -6.79 |
| Covariance contribution | 0.43 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.32 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.92 |
| SVM RNA-class probability | 0.996333 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
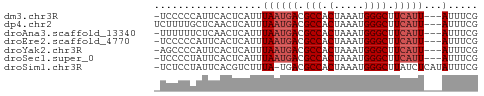
>dm3.chr3R 3987719 50 + 27905053 -UCCCCCAUUCACUCAUUUAAUGACGCCACUAAAUGGGCUUCAUU---AUUUCG -.................((((((.(((.(.....)))).)))))---)..... ( -9.60, z-score = -3.34, R) >dp4.chr2 17711328 51 - 30794189 UCUUUUGCUCAACUCAUUUAAUGACGCCACUAAAUGGGCUUCAUU---AUUUCG ..................((((((.(((.(.....)))).)))))---)..... ( -9.60, z-score = -3.50, R) >droAna3.scaffold_13340 21442312 50 + 23697760 -UUUUUUCUCAACUCAUUUAAUGACGCCACUAAAUGGGCUUCAUU---AUUUCG -.................((((((.(((.(.....)))).)))))---)..... ( -9.60, z-score = -4.50, R) >droEre2.scaffold_4770 153594 50 - 17746568 -UCCCCCAUUCACUCAUUUAAUGACGCCACUAAAUGGGCUUCAUU---AUUUCG -.................((((((.(((.(.....)))).)))))---)..... ( -9.60, z-score = -3.34, R) >droYak2.chr3R 8060585 50 + 28832112 -AGCCCCAUUCACUCAUUUAAUGACGCCACUAAAUGGGCUUCAUU---AUUUCG -.................((((((.(((.(.....)))).)))))---)..... ( -9.60, z-score = -2.67, R) >droSec1.super_0 3279852 50 - 21120651 -UCCCCUAUUCACUCAUUUAAUGACGCCACUAAAUGGGCUUCAUU---AUUUCG -.................((((((.(((.(.....)))).)))))---)..... ( -9.60, z-score = -3.61, R) >droSim1.chr3R 10246465 52 - 27517382 -UCUCCUAUUCACGUCUUUA-UGACGCCACUAAAUGGGCUUAUCUCAUAUUUCG -...((((((..((((....-.))))......))))))................ ( -7.30, z-score = -2.26, R) >consensus _UCCCCCAUUCACUCAUUUAAUGACGCCACUAAAUGGGCUUCAUU___AUUUCG ...................(((((.(((.(.....)))).)))))......... ( -6.36 = -6.79 + 0.43)
| Location | 3,987,719 – 3,987,769 |
|---|---|
| Length | 50 |
| Sequences | 7 |
| Columns | 54 |
| Reading direction | reverse |
| Mean pairwise identity | 84.36 |
| Shannon entropy | 0.29233 |
| G+C content | 0.39663 |
| Mean single sequence MFE | -9.46 |
| Consensus MFE | -7.24 |
| Energy contribution | -7.67 |
| Covariance contribution | 0.43 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.653294 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
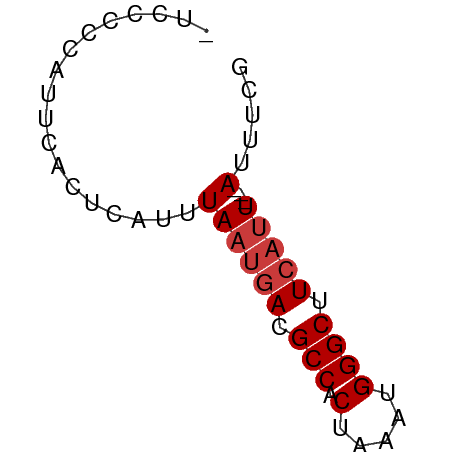
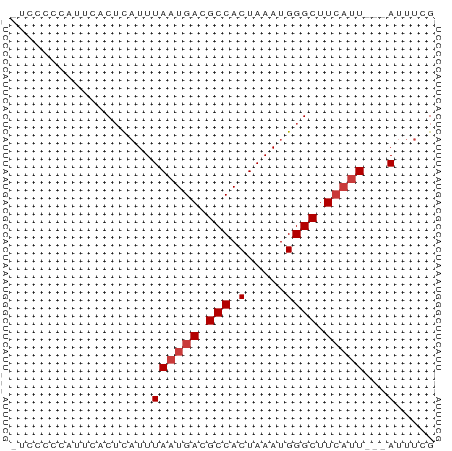
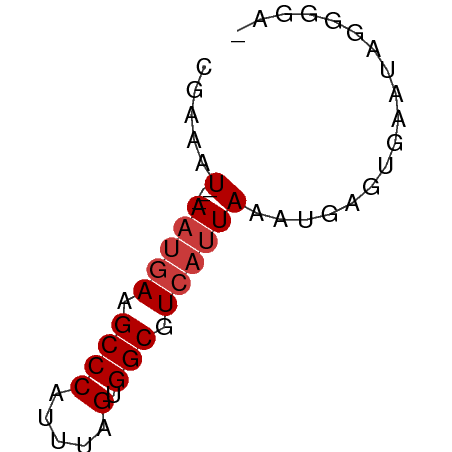
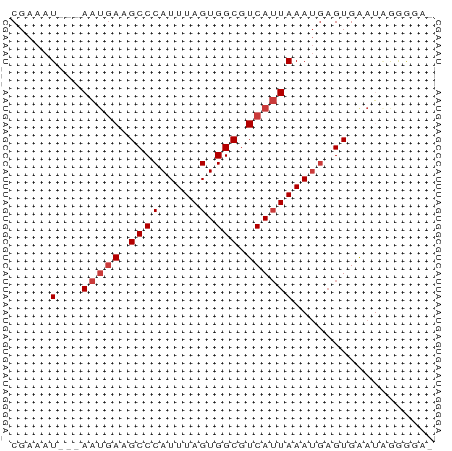
>dm3.chr3R 3987719 50 - 27905053 CGAAAU---AAUGAAGCCCAUUUAGUGGCGUCAUUAAAUGAGUGAAUGGGGGA- ......---.......((((((((....(((......)))..))))))))...- ( -9.50, z-score = -0.90, R) >dp4.chr2 17711328 51 + 30794189 CGAAAU---AAUGAAGCCCAUUUAGUGGCGUCAUUAAAUGAGUUGAGCAAAAGA ......---..((.(((.(((((((((....))))))))).)))...))..... ( -10.30, z-score = -2.29, R) >droAna3.scaffold_13340 21442312 50 - 23697760 CGAAAU---AAUGAAGCCCAUUUAGUGGCGUCAUUAAAUGAGUUGAGAAAAAA- .....(---(((((.((((.....).))).)))))).................- ( -9.40, z-score = -2.60, R) >droEre2.scaffold_4770 153594 50 + 17746568 CGAAAU---AAUGAAGCCCAUUUAGUGGCGUCAUUAAAUGAGUGAAUGGGGGA- ......---.......((((((((....(((......)))..))))))))...- ( -9.50, z-score = -0.90, R) >droYak2.chr3R 8060585 50 - 28832112 CGAAAU---AAUGAAGCCCAUUUAGUGGCGUCAUUAAAUGAGUGAAUGGGGCU- ......---.....(((((((((((((....)))))))).........)))))- ( -10.50, z-score = -1.08, R) >droSec1.super_0 3279852 50 + 21120651 CGAAAU---AAUGAAGCCCAUUUAGUGGCGUCAUUAAAUGAGUGAAUAGGGGA- .....(---(((((.((((.....).))).)))))).................- ( -9.40, z-score = -1.26, R) >droSim1.chr3R 10246465 52 + 27517382 CGAAAUAUGAGAUAAGCCCAUUUAGUGGCGUCA-UAAAGACGUGAAUAGGAGA- ................((.((((....(((((.-....))))))))).))...- ( -7.60, z-score = -1.03, R) >consensus CGAAAU___AAUGAAGCCCAUUUAGUGGCGUCAUUAAAUGAGUGAAUAGGGGA_ ...............((.(((((((((....))))))))).))........... ( -7.24 = -7.67 + 0.43)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:57:42 2011