| Sequence ID | dm3.chr3R |
|---|---|
| Location | 3,981,733 – 3,981,830 |
| Length | 97 |
| Max. P | 0.766796 |

| Location | 3,981,733 – 3,981,830 |
|---|---|
| Length | 97 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 73.19 |
| Shannon entropy | 0.51573 |
| G+C content | 0.48850 |
| Mean single sequence MFE | -29.13 |
| Consensus MFE | -15.16 |
| Energy contribution | -15.07 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.766796 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 3981733 97 + 27905053 ----------UAUUUAUUGCGCAGCUUUUGUGCGCUCUCUUUUU--GAGUCGCCGUUC-AGUGGCAAUGCCCUUUGCCAUCUGAGAUACGAAACGGGACAC----AGUCAAGAU--- ----------........(((((.(....))))))..((((.((--(.(((.((((((-((((((((......)))))).))))........))))))).)----))..)))).--- ( -30.80, z-score = -2.02, R) >droSim1.chr3R 10240308 97 - 27517382 ----------UAUUUAUUGCGCAGCUUUUGUGCGCUCUCUUUUU--GAGUCGCCGUUC-AGUGGCAAUGCCCUUUGCCAUCUGAGAUACGAAACGGGACAC----AGUCAAGAU--- ----------........(((((.(....))))))..((((.((--(.(((.((((((-((((((((......)))))).))))........))))))).)----))..)))).--- ( -30.80, z-score = -2.02, R) >droSec1.super_0 3273886 97 - 21120651 ----------UAUUUAUUGCGCAGCUUUUGUGCGCUCUCUUUUU--GAGUCGCCGUUC-AGUGGCAAUGCCCUUUGCCAUCUGAGAUACGAAACGGGACAC----AGUCAAGAU--- ----------........(((((.(....))))))..((((.((--(.(((.((((((-((((((((......)))))).))))........))))))).)----))..)))).--- ( -30.80, z-score = -2.02, R) >droYak2.chr3R 8054420 97 + 28832112 ----------UAUUUAUUGCGCAGCUUUUGUGCGCUCUCUUUUU--GAGUCGCCGUUC-AGUGGCAAUGCCCUUUGCCAUCUGAGAUACGAGCCGGGACAC----AGUCAAGAU--- ----------........(((((.(....))))))..((((.((--(.(((.((((((-((((((((......)))))).)))))........)))))).)----))..)))).--- ( -29.70, z-score = -1.21, R) >droEre2.scaffold_4770 147575 97 - 17746568 ----------UAUUUAUUGCGCAGCUUUUGUGCGCUCUCUUUUU--GAGUCGCCGUUC-AGUGGCAAUGCCCUUUGCCAUCUGAGAUACGAGCCGGGACAC----AGUCAAGAU--- ----------........(((((.(....))))))..((((.((--(.(((.((((((-((((((((......)))))).)))))........)))))).)----))..)))).--- ( -29.70, z-score = -1.21, R) >droAna3.scaffold_13340 21436781 97 + 23697760 ----------UAUUUAUUGCGCAGCUUUUGUGCGCUCUGCUCUCCAGGGCUUCUGCUCCAGUGGCAAUGCCCUUUGCCAUCUCAGAUA---ACAGGGACAC----AGUCAAGAU--- ----------........(((((.(....)))))).(((...(((..(...((((.....(((((((......)))))))..))))..---.)..)))..)----)).......--- ( -25.80, z-score = 0.55, R) >dp4.chr2 17704263 93 - 30794189 ----------UAUUUAUUGCGCAGCUUUUGUGCGCUCUCUUUUG--A----GCCGGGC-AAUGGCAAUGCCCUUUGCCAUCUCAGAUACGUAACGGGACAC----AGUCAAGAU--- ----------.....(((((.(.((......))((((......)--)----)).).))-)))(((..(((((.((((.(((...)))..)))).))).)).----.))).....--- ( -24.80, z-score = -0.18, R) >droPer1.super_0 7364396 93 + 11822988 ----------UAUUUAUUGCGCAGCUUUUGUGCGCUCUCUUUUG--A----GCCGGGC-AAUGGCAAUGCCCUUUGCCAUCUCAGAUACGCAACGGGACAC----AGUCAAGAU--- ----------.....(((((.(.((......))((((......)--)----)).).))-)))(((..(((((.((((.(((...)))..)))).))).)).----.))).....--- ( -26.80, z-score = -0.59, R) >droWil1.scaffold_181130 14078764 103 + 16660200 CACUCUCGCUUAUUUAUUGCGCAGCUUUUGUGCGCCCUGAAGAU---------GGGGC-AAUGCCAAUGCCCUUUGCCAUCUUAGAUA-GGGUAGAGACGCCCAGAGUUAAGAU--- .((((((((((.......((((((...))))))(((((((((((---------(((((-(.......)))).....)))))))...))-)))).))).))...)))))......--- ( -31.40, z-score = -0.91, R) >droVir3.scaffold_13047 10002471 80 + 19223366 ----------UAUUUAUUGCGCAGCUUUUGUGCGCUGGG--------------------GAUGGCAAUGCCCUUUGCCAUCUUAGAUAUCGCACCAAAGAC--GCAG---GCCAA-- ----------......(((((...((((.(((((...((--------------------((((((((......))))))))))......))))).)))).)--))))---.....-- ( -30.50, z-score = -2.46, R) >droMoj3.scaffold_6540 24788208 85 - 34148556 ----------UAUUUAUUGCGCAGCUUUUGUGCGCUGGG--------------------GAUGGCAAUGCCCUUUGCCAUCUAAGAUAUCGCACCAAAGAC--GCAGCUGGCCAAAA ----------......(((((...((((.(((((.((.(--------------------((((((((......)))))))))....)).))))).)))).)--)))).......... ( -30.60, z-score = -1.80, R) >droGri2.scaffold_14906 6719856 86 - 14172833 ----------UAUUUAUUGCGCAGCUUUUGUGCGCCUUG--------------------GAUGGCAAUGCCCUUUUCCAUCUAAGAUAUCGCACCAAAGGCA-GCACUCAGUCCCAU ----------..........((.(((((((((((.((((--------------------(((((.((......)).)))))))))....)))).))))))).-))............ ( -27.80, z-score = -2.86, R) >consensus __________UAUUUAUUGCGCAGCUUUUGUGCGCUCUCUUUUU__G____GCCGUUC_AAUGGCAAUGCCCUUUGCCAUCUGAGAUACGAAACGGGACAC____AGUCAAGAU___ ..........(((((...(((((.(....)))))).........................(((((((......)))))))...)))))............................. (-15.16 = -15.07 + -0.08)

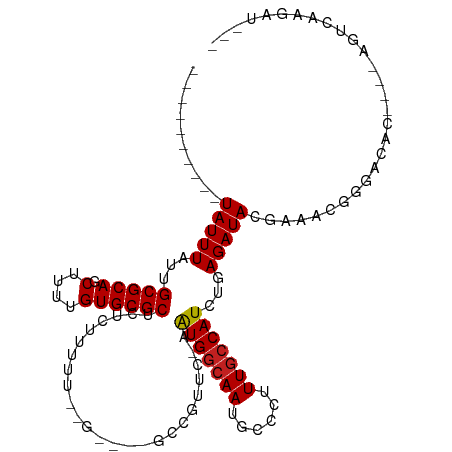

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:57:38 2011