
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 3,966,690 – 3,966,774 |
| Length | 84 |
| Max. P | 0.955277 |

| Location | 3,966,690 – 3,966,774 |
|---|---|
| Length | 84 |
| Sequences | 11 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 84.71 |
| Shannon entropy | 0.33008 |
| G+C content | 0.53852 |
| Mean single sequence MFE | -25.75 |
| Consensus MFE | -24.38 |
| Energy contribution | -23.93 |
| Covariance contribution | -0.45 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.955277 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 3966690 84 + 27905053 CCGAUCCUGCCAGGAGUCGAACCUGGAAUCUUCUGAUCCGUAGUCAGACGCGUUAUCCAUUGCGCCACAGGACCGAUGAGUUUA------------- .((.((((((((((.......))))).....((((((.....)))))).((((........))))..))))).)).........------------- ( -25.60, z-score = -1.55, R) >droSim1.chr3R 10225372 84 - 27517382 CCGAUCCUGCCAGGAGUCGAACCUGGAAUCUUCUGAUCCGUAGUCAGACGCGUUAUCCAUUGCGCCACAGGACCGAUGAAAUUA------------- .((.((((((((((.......))))).....((((((.....)))))).((((........))))..))))).)).........------------- ( -25.60, z-score = -1.92, R) >droSec1.super_0 3258906 84 - 21120651 CCGAUCCUGCCAGGAGUCGAACCUGGAAUCUUCUGAUCCGUAGUCAGACGCGUUAUCCAUUGCGCCACAGGACCGAUGAAAUUA------------- .((.((((((((((.......))))).....((((((.....)))))).((((........))))..))))).)).........------------- ( -25.60, z-score = -1.92, R) >droYak2.chr3R 8039229 84 + 28832112 GCGAUCCUGCCAGGAGUCGAACCUGGAAUCUUCUGAUCCGUAGUCAGACGCGUUAUCCAUUGCGCCACAGGACCGAUGGGUUUA------------- (((......(((((.......))))).....((((((.....)))))))))...((((((((.(((...)).)))))))))...------------- ( -25.60, z-score = -0.63, R) >droEre2.scaffold_4770 132139 84 - 17746568 GCGAUCCUGCCAGGAGUCGAACCUGGAAUCUUCUGAUCCGUAGUCAGACGCGUUAUCCAUUGCGCCACAGGACCGAUGAGCUUA------------- .((.((((((((((.......))))).....((((((.....)))))).((((........))))..))))).)).........------------- ( -25.50, z-score = -1.08, R) >droAna3.scaffold_13340 21421866 77 + 23697760 UCGAUCCUGCCAGGAGUCGAACCUGGAAUCUUCUGAUCCGUAGUCAGACGCGUUAUCCAUUGCGCCACAGGACCGGA-------------------- (((.((((((((((.......))))).....((((((.....)))))).((((........))))..))))).))).-------------------- ( -26.00, z-score = -1.54, R) >droWil1.scaffold_180700 756566 97 - 6630534 GCGAUCCUGCCAGGAGUCGAACCUGGAAUCUUCUGAUCCGUAGUCAGACGCGUUAUCCAUUGCGCCACAGGACCGAUAUGAAAUAUGUUGGCUAAGA ....((((((((((.......))))).....((((((.....)))))).((((........))))..)))))((((((((...))))))))...... ( -28.40, z-score = -1.16, R) >droMoj3.scaffold_6496 25293158 84 + 26866924 GCGAUUCUGCCAGGAGUCGAACCUGGAAUCUUCUGAUCCGUAGUCAGACGCGUUAUCCAUUGCGCCACAGGACCAAUGAAUCGG------------- .((((((..(((((.......)))))..(((((((((.....)))))).((((........))))....))).....)))))).------------- ( -24.50, z-score = -0.91, R) >droVir3.scaffold_12875 15834067 84 - 20611582 ACGAUCCUGCCAGGAGUCGAACCUGGAAUCUUCUGAUCCGUAGUCAGACGCGUUAUCCAUUGCGCCACAGGACCGACUGAUCGU------------- ((((((((....))(((((..((((......((((((.....)))))).((((........))))..))))..)))))))))))------------- ( -27.90, z-score = -1.80, R) >droGri2.scaffold_15245 208877 84 + 18325388 UCGAUCCUGCCAGGAGUCGAACCUGGAAUCUUCUGAUCCGUAGUCAGACGCGUUAUCCAUUGCGCCACAGGACCGAUUGAUCAG------------- (((.((((((((((.......))))).....((((((.....)))))).((((........))))..))))).)))........------------- ( -26.70, z-score = -1.76, R) >anoGam1.chr2R 57351005 80 + 62725911 --GACCCCGACGUGAUUUGAACACGCAACCUUCUGAUCUGGAGUCAGACGCGCUA-CCGUUGCGCCACGGAGUCCCAUACUUU-------------- --(((.(((.((((.......))))......((((((.....)))))).((((..-.....))))..))).))).........-------------- ( -21.80, z-score = -1.60, R) >consensus GCGAUCCUGCCAGGAGUCGAACCUGGAAUCUUCUGAUCCGUAGUCAGACGCGUUAUCCAUUGCGCCACAGGACCGAUGAAUUUA_____________ .((.((((((((((.......))))).....((((((.....)))))).((((........))))..))))).))...................... (-24.38 = -23.93 + -0.45)

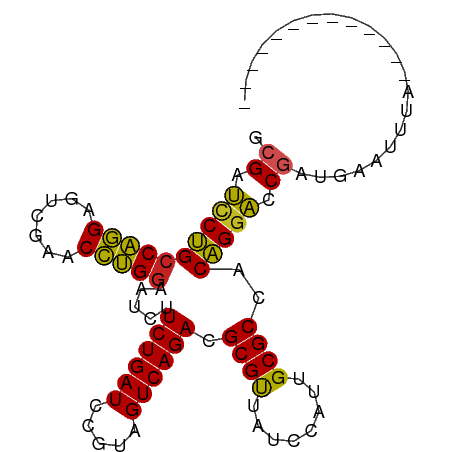
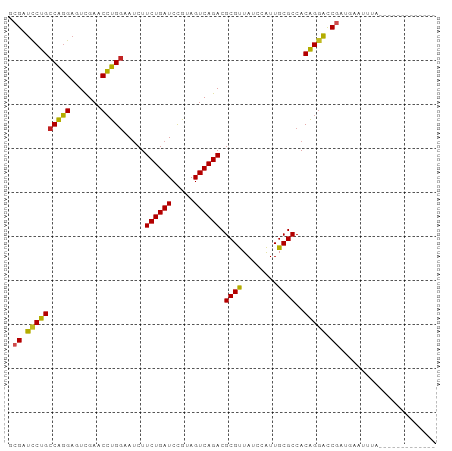
| Location | 3,966,690 – 3,966,774 |
|---|---|
| Length | 84 |
| Sequences | 11 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 84.71 |
| Shannon entropy | 0.33008 |
| G+C content | 0.53852 |
| Mean single sequence MFE | -28.01 |
| Consensus MFE | -27.20 |
| Energy contribution | -26.33 |
| Covariance contribution | -0.87 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.97 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.48 |
| SVM RNA-class probability | 0.942519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 3966690 84 - 27905053 -------------UAAACUCAUCGGUCCUGUGGCGCAAUGGAUAACGCGUCUGACUACGGAUCAGAAGAUUCCAGGUUCGACUCCUGGCAGGAUCGG -------------.........((((((((((((((..........))))).......(((((....)))))((((.......))))))))))))). ( -28.70, z-score = -1.78, R) >droSim1.chr3R 10225372 84 + 27517382 -------------UAAUUUCAUCGGUCCUGUGGCGCAAUGGAUAACGCGUCUGACUACGGAUCAGAAGAUUCCAGGUUCGACUCCUGGCAGGAUCGG -------------.........((((((((((((((..........))))).......(((((....)))))((((.......))))))))))))). ( -28.70, z-score = -1.78, R) >droSec1.super_0 3258906 84 + 21120651 -------------UAAUUUCAUCGGUCCUGUGGCGCAAUGGAUAACGCGUCUGACUACGGAUCAGAAGAUUCCAGGUUCGACUCCUGGCAGGAUCGG -------------.........((((((((((((((..........))))).......(((((....)))))((((.......))))))))))))). ( -28.70, z-score = -1.78, R) >droYak2.chr3R 8039229 84 - 28832112 -------------UAAACCCAUCGGUCCUGUGGCGCAAUGGAUAACGCGUCUGACUACGGAUCAGAAGAUUCCAGGUUCGACUCCUGGCAGGAUCGC -------------.........((((((((((((((..........))))).......(((((....)))))((((.......))))))))))))). ( -27.80, z-score = -1.32, R) >droEre2.scaffold_4770 132139 84 + 17746568 -------------UAAGCUCAUCGGUCCUGUGGCGCAAUGGAUAACGCGUCUGACUACGGAUCAGAAGAUUCCAGGUUCGACUCCUGGCAGGAUCGC -------------.........((((((((((((((..........))))).......(((((....)))))((((.......))))))))))))). ( -27.80, z-score = -1.09, R) >droAna3.scaffold_13340 21421866 77 - 23697760 --------------------UCCGGUCCUGUGGCGCAAUGGAUAACGCGUCUGACUACGGAUCAGAAGAUUCCAGGUUCGACUCCUGGCAGGAUCGA --------------------..((((((((((((((..........))))).......(((((....)))))((((.......))))))))))))). ( -28.70, z-score = -1.57, R) >droWil1.scaffold_180700 756566 97 + 6630534 UCUUAGCCAACAUAUUUCAUAUCGGUCCUGUGGCGCAAUGGAUAACGCGUCUGACUACGGAUCAGAAGAUUCCAGGUUCGACUCCUGGCAGGAUCGC ......................((((((((((((((..........))))).......(((((....)))))((((.......))))))))))))). ( -27.80, z-score = -0.78, R) >droMoj3.scaffold_6496 25293158 84 - 26866924 -------------CCGAUUCAUUGGUCCUGUGGCGCAAUGGAUAACGCGUCUGACUACGGAUCAGAAGAUUCCAGGUUCGACUCCUGGCAGAAUCGC -------------.((((((.(((((((.(((((((..........)))))..))...)))))))......(((((.......)))))..)))))). ( -27.70, z-score = -1.30, R) >droVir3.scaffold_12875 15834067 84 + 20611582 -------------ACGAUCAGUCGGUCCUGUGGCGCAAUGGAUAACGCGUCUGACUACGGAUCAGAAGAUUCCAGGUUCGACUCCUGGCAGGAUCGU -------------.........((((((((((((((..........))))).......(((((....)))))((((.......))))))))))))). ( -28.00, z-score = -0.66, R) >droGri2.scaffold_15245 208877 84 - 18325388 -------------CUGAUCAAUCGGUCCUGUGGCGCAAUGGAUAACGCGUCUGACUACGGAUCAGAAGAUUCCAGGUUCGACUCCUGGCAGGAUCGA -------------........(((((((((((((((..........))))).......(((((....)))))((((.......)))))))))))))) ( -29.50, z-score = -1.48, R) >anoGam1.chr2R 57351005 80 - 62725911 --------------AAAGUAUGGGACUCCGUGGCGCAACGG-UAGCGCGUCUGACUCCAGAUCAGAAGGUUGCGUGUUCAAAUCACGUCGGGGUC-- --------------.........(((((((..((((.....-..)))).((((((....).))))).....(((((.......))))))))))))-- ( -24.70, z-score = -0.53, R) >consensus _____________UAAAUUCAUCGGUCCUGUGGCGCAAUGGAUAACGCGUCUGACUACGGAUCAGAAGAUUCCAGGUUCGACUCCUGGCAGGAUCGC ......................((((((((..(((..........))).((((((....).))))).....(((((.......))))))))))))). (-27.20 = -26.33 + -0.87)

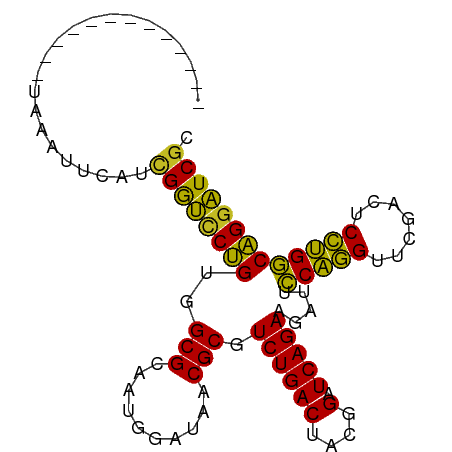
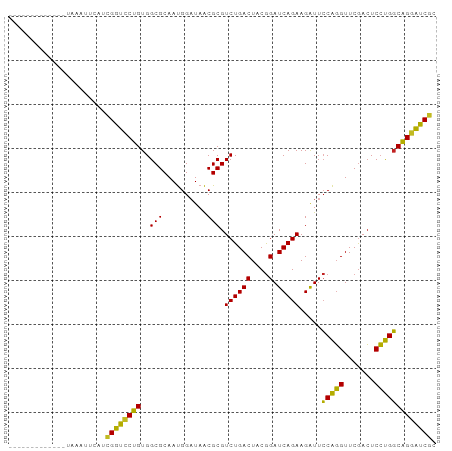
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:57:36 2011