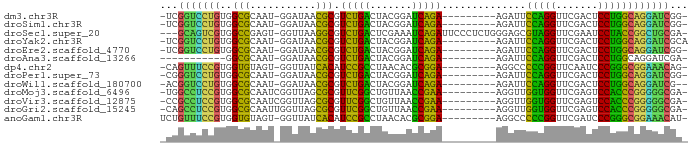
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 3,966,447 – 3,966,523 |
| Length | 76 |
| Max. P | 0.997761 |

| Location | 3,966,447 – 3,966,523 |
|---|---|
| Length | 76 |
| Sequences | 13 |
| Columns | 88 |
| Reading direction | forward |
| Mean pairwise identity | 68.63 |
| Shannon entropy | 0.63413 |
| G+C content | 0.58493 |
| Mean single sequence MFE | -27.02 |
| Consensus MFE | -24.23 |
| Energy contribution | -20.90 |
| Covariance contribution | -3.33 |
| Combinations/Pair | 1.85 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.17 |
| SVM RNA-class probability | 0.997761 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
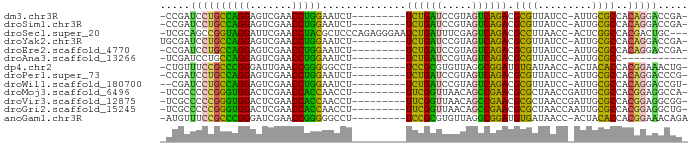
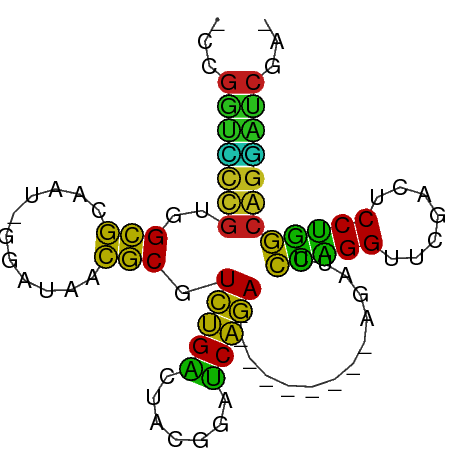
>dm3.chr3R 3966447 76 + 27905053 -CCGAUCCUGCCAGGAGUCGAACCUGGAAUCU---------UCUGAUCCGUAGUCAGACGCGUUAUCC-AUUGCGCCACAGGACCGA- -.((.((((((((((.......))))).....---------((((((.....)))))).((((.....-...))))..))))).)).- ( -25.60, z-score = -2.08, R) >droSim1.chr3R 10225134 76 - 27517382 -CCGAUCCUGCCAGGAGUCGAACCUGGAAUCU---------UCUGAUCCGUAGUCAGACGCGUUAUCC-AUUGCGCCACAGGACCGA- -.((.((((((((((.......))))).....---------((((((.....)))))).((((.....-...))))..))))).)).- ( -25.60, z-score = -2.08, R) >droSec1.super_20 605364 83 - 1148123 -UCGCAGCCGGUAGGAUUCGAACCUACGCUCCCAGAGGGAAUCUGAUUUCGAGUCAGACGCCUUAACC-ACUCGGCCACGACUGC--- -(((..(((((((((.......)))))........((((..((((((.....))))))..))))....-...))))..)))....--- ( -27.00, z-score = -1.57, R) >droYak2.chr3R 8038966 77 + 28832112 UGCGAUCCUGCCAGGAGUCGAACCUGGAAUCU---------UCUGAUCCGUAGUCAGACGCGUUAUCC-AUUGCGCCACAGGACCGA- ..((.((((((((((.......))))).....---------((((((.....)))))).((((.....-...))))..))))).)).- ( -25.50, z-score = -1.70, R) >droEre2.scaffold_4770 131891 76 - 17746568 -CCGAUCCUGCCAGGAGUCGAACCUGGAAUCU---------UCUGAUCCGUAGUCAGACGCGUUAUCC-AUUGCGCCACAGGACCGA- -.((.((((((((((.......))))).....---------((((((.....)))))).((((.....-...))))..))))).)).- ( -25.60, z-score = -2.08, R) >droAna3.scaffold_13266 18744044 66 + 19884421 -UCGAUCCUGCCAGGAGUCGAACCUGGAAUCU---------UCUGAUCCGUAGUCAGACGCGUUAUCC-AUUGCGCC----------- -..(((....(((((.......))))).))).---------((((((.....)))))).((((.....-...)))).----------- ( -17.90, z-score = -0.84, R) >dp4.chr2 7051452 76 - 30794189 -CUGUUUCCGCCCGGGAUUGAACCGGGGGCCU---------UCCGCGUGUUAGGCGGAUGUGAUAACC-ACUACACCACGGAAACUG- -..((((((((((((.......)))))((...---------(((((.......))))).(((.....)-))....)).)))))))..- ( -30.10, z-score = -2.16, R) >droPer1.super_73 202111 76 - 267513 -CCGAUCCUGCCAGGAGUCGAACCUGGAAUCU---------UCUGAUCCGUAGUCAGACGCGUUAUCC-AUUGCGCCACAGGACCCG- -....((((((((((.......))))).....---------((((((.....)))))).((((.....-...))))..)))))....- ( -23.20, z-score = -1.66, R) >droWil1.scaffold_180700 738705 75 - 6630534 --CGAUCCUGCCAGGAGUCGAACCUGGAAUCU---------UCUGAUCCGUAGUCAGACGCGUUAUCC-AUUGCGCCACAGGACCGU- --((.((((((((((.......))))).....---------((((((.....)))))).((((.....-...))))..))))).)).- ( -24.80, z-score = -1.82, R) >droMoj3.scaffold_6496 25292866 77 + 26866924 -UCGCCCCCGGGUGGACUCGAACCACCAACCU---------UUCGGUUAACAGCCGAACGCGCUAACCGAUUGCGCCACGGAGGCCA- -..(((.((((((((.......))))).....---------((((((.....)))))).((((.........))))..))).)))..- ( -30.40, z-score = -2.55, R) >droVir3.scaffold_12875 15833733 77 - 20611582 -UCGCCCCCGGGUGGACUCGAACCACCAACCU---------UUCGGUUAACAGCCGAACGCGCUAACCGAUUGCGCCACGGAGGCGG- -.((((.((((((((.......))))).....---------((((((.....)))))).((((.........))))..))).)))).- ( -33.10, z-score = -2.88, R) >droGri2.scaffold_15245 208595 77 + 18325388 -UCGCCCCCGGGUGGACUCGAACCACCAACCU---------UUCGGUUAACAGCCGAACGCGCUAACCAAUUGCGCCACGGAGGCUG- -..(((.((((((((.......))))).....---------((((((.....)))))).((((.........))))..))).)))..- ( -30.80, z-score = -3.21, R) >anoGam1.chr3R 13656675 77 + 53272125 -AUGUUUCCGCCCGGGAUCGAACCGGGGGCCU---------UCCGCGUGUUAGGCGGAUGUGAUAACC-ACUACACCACGGAAACAGA -.(((((((((((((.......)))))((...---------(((((.......))))).(((.....)-))....)).)))))))).. ( -31.60, z-score = -2.12, R) >consensus _CCGAUCCUGCCAGGAGUCGAACCUGGAAUCU_________UCUGAUCCGUAGUCAGACGCGUUAUCC_AUUGCGCCACAGGACCGA_ .....((((((((((.......)))))..............((((((.....)))))).((((.........))))..)))))..... (-24.23 = -20.90 + -3.33)
| Location | 3,966,447 – 3,966,523 |
|---|---|
| Length | 76 |
| Sequences | 13 |
| Columns | 88 |
| Reading direction | reverse |
| Mean pairwise identity | 68.63 |
| Shannon entropy | 0.63413 |
| G+C content | 0.58493 |
| Mean single sequence MFE | -30.09 |
| Consensus MFE | -27.53 |
| Energy contribution | -23.03 |
| Covariance contribution | -4.50 |
| Combinations/Pair | 2.00 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.93 |
| SVM RNA-class probability | 0.996458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 3966447 76 - 27905053 -UCGGUCCUGUGGCGCAAU-GGAUAACGCGUCUGACUACGGAUCAGA---------AGAUUCCAGGUUCGACUCCUGGCAGGAUCGG- -.((((((((((((((...-.......))))).......(((((...---------.)))))((((.......))))))))))))).- ( -28.70, z-score = -1.86, R) >droSim1.chr3R 10225134 76 + 27517382 -UCGGUCCUGUGGCGCAAU-GGAUAACGCGUCUGACUACGGAUCAGA---------AGAUUCCAGGUUCGACUCCUGGCAGGAUCGG- -.((((((((((((((...-.......))))).......(((((...---------.)))))((((.......))))))))))))).- ( -28.70, z-score = -1.86, R) >droSec1.super_20 605364 83 + 1148123 ---GCAGUCGUGGCCGAGU-GGUUAAGGCGUCUGACUCGAAAUCAGAUUCCCUCUGGGAGCGUAGGUUCGAAUCCUACCGGCUGCGA- ---((((((((((((....-)))))..((((((((.......))))))((((...))))))(((((.......))))))))))))..- ( -31.20, z-score = -1.27, R) >droYak2.chr3R 8038966 77 - 28832112 -UCGGUCCUGUGGCGCAAU-GGAUAACGCGUCUGACUACGGAUCAGA---------AGAUUCCAGGUUCGACUCCUGGCAGGAUCGCA -.((((((((((((((...-.......))))).......(((((...---------.)))))((((.......))))))))))))).. ( -27.80, z-score = -1.44, R) >droEre2.scaffold_4770 131891 76 + 17746568 -UCGGUCCUGUGGCGCAAU-GGAUAACGCGUCUGACUACGGAUCAGA---------AGAUUCCAGGUUCGACUCCUGGCAGGAUCGG- -.((((((((((((((...-.......))))).......(((((...---------.)))))((((.......))))))))))))).- ( -28.70, z-score = -1.86, R) >droAna3.scaffold_13266 18744044 66 - 19884421 -----------GGCGCAAU-GGAUAACGCGUCUGACUACGGAUCAGA---------AGAUUCCAGGUUCGACUCCUGGCAGGAUCGA- -----------.(((....-......))).((((((....).)))))---------.(((((((((.......)))))...))))..- ( -18.10, z-score = -0.16, R) >dp4.chr2 7051452 76 + 30794189 -CAGUUUCCGUGGUGUAGU-GGUUAUCACAUCCGCCUAACACGCGGA---------AGGCCCCCGGUUCAAUCCCGGGCGGAAACAG- -..(((((((.(((.(.((-(.....))).(((((.......)))))---------).)))(((((.......))))))))))))..- ( -30.10, z-score = -2.51, R) >droPer1.super_73 202111 76 + 267513 -CGGGUCCUGUGGCGCAAU-GGAUAACGCGUCUGACUACGGAUCAGA---------AGAUUCCAGGUUCGACUCCUGGCAGGAUCGG- -..(((((((((((((...-.......))))).......(((((...---------.)))))((((.......))))))))))))..- ( -26.60, z-score = -1.09, R) >droWil1.scaffold_180700 738705 75 + 6630534 -ACGGUCCUGUGGCGCAAU-GGAUAACGCGUCUGACUACGGAUCAGA---------AGAUUCCAGGUUCGACUCCUGGCAGGAUCG-- -.((((((((((((((...-.......))))).......(((((...---------.)))))((((.......)))))))))))))-- ( -27.80, z-score = -1.89, R) >droMoj3.scaffold_6496 25292866 77 - 26866924 -UGGCCUCCGUGGCGCAAUCGGUUAGCGCGUUCGGCUGUUAACCGAA---------AGGUUGGUGGUUCGAGUCCACCCGGGGGCGA- -..(((((((.(((((.........)))).((((((..((((((...---------.))))))..).)))))....).)))))))..- ( -36.90, z-score = -2.27, R) >droVir3.scaffold_12875 15833733 77 + 20611582 -CCGCCUCCGUGGCGCAAUCGGUUAGCGCGUUCGGCUGUUAACCGAA---------AGGUUGGUGGUUCGAGUCCACCCGGGGGCGA- -.((((((((.(((((.........)))).((((((..((((((...---------.))))))..).)))))....).)))))))).- ( -39.00, z-score = -2.78, R) >droGri2.scaffold_15245 208595 77 - 18325388 -CAGCCUCCGUGGCGCAAUUGGUUAGCGCGUUCGGCUGUUAACCGAA---------AGGUUGGUGGUUCGAGUCCACCCGGGGGCGA- -..(((((((.(((((.........)))).((((((..((((((...---------.))))))..).)))))....).)))))))..- ( -37.10, z-score = -2.89, R) >anoGam1.chr3R 13656675 77 - 53272125 UCUGUUUCCGUGGUGUAGU-GGUUAUCACAUCCGCCUAACACGCGGA---------AGGCCCCCGGUUCGAUCCCGGGCGGAAACAU- ..((((((((.(((.(.((-(.....))).(((((.......)))))---------).)))(((((.......))))))))))))).- ( -30.50, z-score = -1.87, R) >consensus _CCGGUCCCGUGGCGCAAU_GGAUAACGCGUCUGACUACGGAUCAGA_________AGAUUCCAGGUUCGACUCCUGGCAGGAUCGA_ ...(((((((..(((...........))).(((((.......)))))..............(((((.......))))))))))))... (-27.53 = -23.03 + -4.50)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:57:34 2011