
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 3,892,231 – 3,892,326 |
| Length | 95 |
| Max. P | 0.999252 |

| Location | 3,892,231 – 3,892,326 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 69.28 |
| Shannon entropy | 0.54235 |
| G+C content | 0.63195 |
| Mean single sequence MFE | -29.87 |
| Consensus MFE | -19.55 |
| Energy contribution | -19.61 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.70 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.71 |
| SVM RNA-class probability | 0.999213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
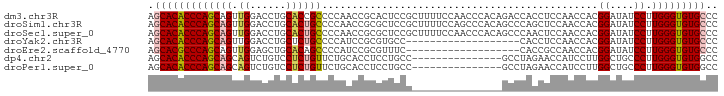
>dm3.chr3R 3892231 95 + 27905053 AGCACACCCAGCAGUUGGACCUGCACCGCCCCAACCGCACUCCGCUUUUCCAACCCACAGACCACCUCCAACCACGGAUAUCCUUGGGUGUGCCC .((((((((((..((((((...((...((.......)).....))...))))))............(((......))).....)))))))))).. ( -28.20, z-score = -3.23, R) >droSim1.chr3R 10159869 95 - 27517382 AGCACACCCAGCAGUUGGACCUGCACUGCCCCAACCGCGCUCCGCUUUUCCAGCCCACAGCCCAGCUCCAACCACGGAUAUCCUUGGGUGUGCCC .((((((((((..((((((.(((....((.......))(((..(((.....)))....))).))).))))))...((....)))))))))))).. ( -33.20, z-score = -3.02, R) >droSec1.super_0 3195923 95 - 21120651 AGCACACCCAGCAGUUGGACCUGCACUGCCCCAACCGCGCUCCGCUUUUCCAACCCACAGCCCAACUCCAACCACGGAUAUCCUUGGGUGUGCCC .((((((((((..((((((...((...(((......).))...))...))))))............(((......))).....)))))))))).. ( -30.20, z-score = -3.07, R) >droYak2.chr3R 7971983 76 + 28832112 AGCACACCCAGCAGUUGGACCUGCUCUGCCCCAUCCGCGUGCC-------------------CACCUCCAACCACGGAUAUCCUUGGGUGUGCCC .((((((((((..((((((..((..((((.......))).)..-------------------))..))))))...((....)))))))))))).. ( -28.00, z-score = -2.76, R) >droEre2.scaffold_4770 66370 76 - 17746568 AGCACGCCCAGCAGUUGGAGCUGCACAGCCCCAUCCGCGUUUC-------------------CACCGCCAACCACGGAUAUCCUUGGGUGUGCCC .(((((((((((.(.(((.(((....))).))).).)).....-------------------..(((.......))).......))))))))).. ( -29.80, z-score = -2.96, R) >dp4.chr2 24396963 80 - 30794189 AGCACACCCAGCAGCAGUCUGUCCUCUGUUCUGCACCUCCUGCC---------------GCCUAGAACCAUCCUUGGCUGCCCUUGGGUGUGGCC ..(((((((((..((((((........((((((((.....))).---------------....))))).......))))))..)))))))))... ( -29.86, z-score = -1.93, R) >droPer1.super_0 11602153 80 - 11822988 AGCACACCCAGCAGCAGUCUGUCCUCUGUUCUGCACCUCCUGCC---------------GCCUAGAACCAUCCUUGGCUGCCCUUGGGUGUGGCC ..(((((((((..((((((........((((((((.....))).---------------....))))).......))))))..)))))))))... ( -29.86, z-score = -1.93, R) >consensus AGCACACCCAGCAGUUGGACCUGCACUGCCCCAACCGCGCUCCG_______________GCCCACCUCCAACCACGGAUAUCCUUGGGUGUGCCC .(((((((((((((.((......))))))..............................................((....)).))))))))).. (-19.55 = -19.61 + 0.06)

| Location | 3,892,231 – 3,892,326 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 69.28 |
| Shannon entropy | 0.54235 |
| G+C content | 0.63195 |
| Mean single sequence MFE | -40.56 |
| Consensus MFE | -21.79 |
| Energy contribution | -21.77 |
| Covariance contribution | -0.02 |
| Combinations/Pair | 1.25 |
| Mean z-score | -3.02 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.74 |
| SVM RNA-class probability | 0.999252 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
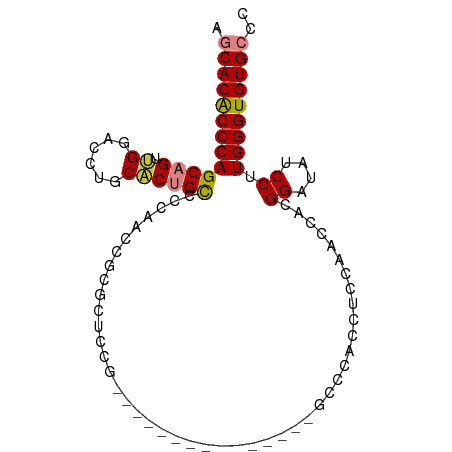
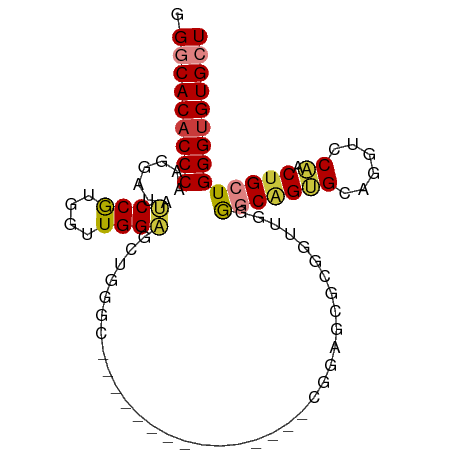
>dm3.chr3R 3892231 95 - 27905053 GGGCACACCCAAGGAUAUCCGUGGUUGGAGGUGGUCUGUGGGUUGGAAAAGCGGAGUGCGGUUGGGGCGGUGCAGGUCCAACUGCUGGGUGUGCU .((((((((((.((....))(..((((((.....(((((...........)))))(..(.((....)).)..)...))))))..))))))))))) ( -41.70, z-score = -2.93, R) >droSim1.chr3R 10159869 95 + 27517382 GGGCACACCCAAGGAUAUCCGUGGUUGGAGCUGGGCUGUGGGCUGGAAAAGCGGAGCGCGGUUGGGGCAGUGCAGGUCCAACUGCUGGGUGUGCU .((((((((((.((....))(..((((((.(((.(((((...((.((...(((...)))..)).))))))).))).))))))..))))))))))) ( -51.80, z-score = -4.70, R) >droSec1.super_0 3195923 95 + 21120651 GGGCACACCCAAGGAUAUCCGUGGUUGGAGUUGGGCUGUGGGUUGGAAAAGCGGAGCGCGGUUGGGGCAGUGCAGGUCCAACUGCUGGGUGUGCU .((((((((((.((....))(..((((((.(((.(((((...(..(....(((...)))..)..).))))).))).))))))..))))))))))) ( -47.70, z-score = -4.40, R) >droYak2.chr3R 7971983 76 - 28832112 GGGCACACCCAAGGAUAUCCGUGGUUGGAGGUG-------------------GGCACGCGGAUGGGGCAGAGCAGGUCCAACUGCUGGGUGUGCU .(((((((((.....(((((((((((.......-------------------))).))))))))......(((((......)))))))))))))) ( -36.60, z-score = -2.91, R) >droEre2.scaffold_4770 66370 76 + 17746568 GGGCACACCCAAGGAUAUCCGUGGUUGGCGGUG-------------------GAAACGCGGAUGGGGCUGUGCAGCUCCAACUGCUGGGCGUGCU .(((((.((((.((....))(..(((..(.(((-------------------....))).)..((((((....)))))))))..))))).))))) ( -37.30, z-score = -2.61, R) >dp4.chr2 24396963 80 + 30794189 GGCCACACCCAAGGGCAGCCAAGGAUGGUUCUAGGC---------------GGCAGGAGGUGCAGAACAGAGGACAGACUGCUGCUGGGUGUGCU (((.((((((...((.(((((....))))))).(((---------------(((((...((.(........).))...))))))))))))))))) ( -34.40, z-score = -1.81, R) >droPer1.super_0 11602153 80 + 11822988 GGCCACACCCAAGGGCAGCCAAGGAUGGUUCUAGGC---------------GGCAGGAGGUGCAGAACAGAGGACAGACUGCUGCUGGGUGUGCU (((.((((((...((.(((((....))))))).(((---------------(((((...((.(........).))...))))))))))))))))) ( -34.40, z-score = -1.81, R) >consensus GGGCACACCCAAGGAUAUCCGUGGUUGGAGCUGGGC_______________CGGAGCGCGGUUGGGGCAGUGCAGGUCCAACUGCUGGGUGUGCU .(((((((((.......((((....))))....................................(((((((......)).)))))))))))))) (-21.79 = -21.77 + -0.02)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:57:22 2011