
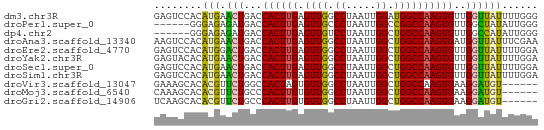
| Sequence ID | dm3.chr3R |
|---|---|
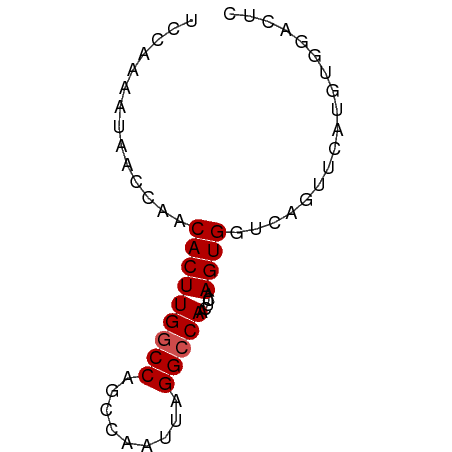
| Location | 3,889,295 – 3,889,359 |
| Length | 64 |
| Max. P | 0.997040 |

| Location | 3,889,295 – 3,889,359 |
|---|---|
| Length | 64 |
| Sequences | 11 |
| Columns | 64 |
| Reading direction | forward |
| Mean pairwise identity | 77.77 |
| Shannon entropy | 0.48343 |
| G+C content | 0.49094 |
| Mean single sequence MFE | -15.22 |
| Consensus MFE | -8.92 |
| Energy contribution | -9.10 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.19 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.689703 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
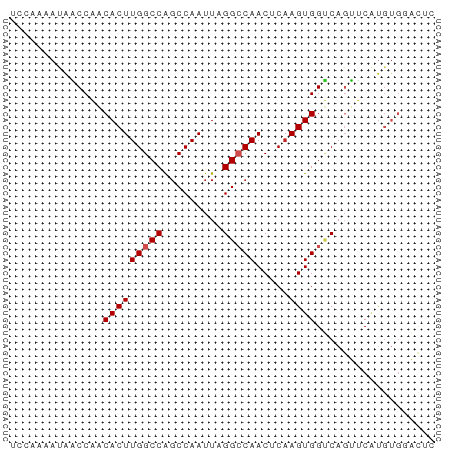
>dm3.chr3R 3889295 64 + 27905053 CCCAAAAUAACCAACACUUGGCCAUCCAAUUAGGCCAACUCAAGUGGUCAGUUCAUGUGGACUC .........((((....((((((.........))))))......)))).(((((....))))). ( -15.00, z-score = -1.35, R) >droPer1.super_0 11599551 58 - 11822988 CCCAAUAUAGCCAACACUUGGCCGGCCAAUUAGGGCAACUCAAGUGGUCAUCUCUCCC------ .........((((.....)))).(((((.((..(....)..)).))))).........------ ( -15.10, z-score = -1.40, R) >dp4.chr2 24394337 58 - 30794189 CCCAAUAUGGCCAACACUUGGCCAGCCAAUUAGGACAACUCAAGUGGUCAUCUCUCCC------ .......((((((.....))))))((((...((.....))....))))..........------ ( -13.20, z-score = -0.92, R) >droAna3.scaffold_13340 15160176 64 - 23697760 UUCGAAAUAACCAUCACUUGGCCAGCCAAUUAGGCCAACUCAAGUGGUCAGUUCAUGUGGACUU ............(((((((((...(((.....)))....))))))))).(((((....))))). ( -16.10, z-score = -1.46, R) >droEre2.scaffold_4770 63484 64 - 17746568 UCCAAAAUAACCAACACUUGGCCAGCCAAUUAGGCCAACUCAAGUGGUCAGUCCAUGUGGACUC ..............(((((((...(((.....)))....)))))))...(((((....))))). ( -18.00, z-score = -2.33, R) >droYak2.chr3R 7968946 64 + 28832112 UCCAAAAUAACCAACACUUGGCCAGCCAAUUAGGCCAACUCAAGUGGUCAGUUCAUGUGUACUC .............((((((((((.........)))))).....((((.....)))))))).... ( -12.80, z-score = -0.95, R) >droSec1.super_0 3193102 64 - 21120651 UCCAAAAUAACCAACACUUGGCCAGCCAAUUAGGCCAACUCAAGUGGUCAGUUCAUGUGGACUC ((((....(((...(((((((...(((.....)))....)))))))....)))....))))... ( -16.80, z-score = -1.93, R) >droSim1.chr3R 10156986 64 - 27517382 UCCAAAAUAACCAACACUUGGCCAGCCAAUUAGGCCAACUCAAGUGGUCAGUUCAUGUGGACUC ((((....(((...(((((((...(((.....)))....)))))))....)))....))))... ( -16.80, z-score = -1.93, R) >droVir3.scaffold_13047 9912615 58 + 19223366 ------ACAUCCUUCACUUGGCCAGCCAAUUAGGCCAACACUAGUGGCCAGAACGUGUGCUUUC ------((((..(((..((((....))))...(((((.......))))).))).))))...... ( -14.20, z-score = -0.67, R) >droMoj3.scaffold_6540 24685519 58 - 34148556 ------ACAUCCUUCACUUGGCCAGCCAAUUAGGCCAACACAAGUGGGCAGAACGUGUGCUUUG ------........((((((....(((.....))).....))))))((((.......))))... ( -13.50, z-score = 0.25, R) >droGri2.scaffold_14906 6639300 58 - 14172833 ------ACAUCCUUCACUUGGCCAGCCAAUUAGGCCAACACAAGUGGGCAGAACGUGUGCUUGA ------...........((((((.........))))))..((((((.((.....)).)))))). ( -15.90, z-score = -0.36, R) >consensus UCCAAAAUAACCAACACUUGGCCAGCCAAUUAGGCCAACUCAAGUGGUCAGUUCAUGUGGACUC ..............(((((((((.........))))).....)))).................. ( -8.92 = -9.10 + 0.18)

| Location | 3,889,295 – 3,889,359 |
|---|---|
| Length | 64 |
| Sequences | 11 |
| Columns | 64 |
| Reading direction | reverse |
| Mean pairwise identity | 77.77 |
| Shannon entropy | 0.48343 |
| G+C content | 0.49094 |
| Mean single sequence MFE | -23.43 |
| Consensus MFE | -11.68 |
| Energy contribution | -11.27 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.20 |
| Mean z-score | -3.00 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.03 |
| SVM RNA-class probability | 0.997040 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
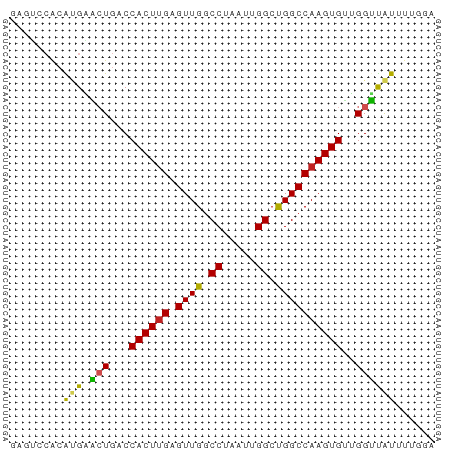
>dm3.chr3R 3889295 64 - 27905053 GAGUCCACAUGAACUGACCACUUGAGUUGGCCUAAUUGGAUGGCCAAGUGUUGGUUAUUUUGGG ....(((....(((..((.(((((.(((..((.....))..))))))))))..)))....))). ( -19.80, z-score = -1.66, R) >droPer1.super_0 11599551 58 + 11822988 ------GGGAGAGAUGACCACUUGAGUUGCCCUAAUUGGCCGGCCAAGUGUUGGCUAUAUUGGG ------((.........))...........((((((((((((((.....)))))))).)))))) ( -16.20, z-score = -0.41, R) >dp4.chr2 24394337 58 + 30794189 ------GGGAGAGAUGACCACUUGAGUUGUCCUAAUUGGCUGGCCAAGUGUUGGCCAUAUUGGG ------......((..(((....).))..))(((((((((..((.....))..)))).))))). ( -17.60, z-score = -0.92, R) >droAna3.scaffold_13340 15160176 64 + 23697760 AAGUCCACAUGAACUGACCACUUGAGUUGGCCUAAUUGGCUGGCCAAGUGAUGGUUAUUUCGAA .........((((.((((((((((.((..(((.....)))..)))))))...))))).)))).. ( -23.60, z-score = -3.61, R) >droEre2.scaffold_4770 63484 64 + 17746568 GAGUCCACAUGGACUGACCACUUGAGUUGGCCUAAUUGGCUGGCCAAGUGUUGGUUAUUUUGGA ...((((....(((..((.(((((.((..(((.....)))..)))))))))..)))....)))) ( -28.30, z-score = -4.34, R) >droYak2.chr3R 7968946 64 - 28832112 GAGUACACAUGAACUGACCACUUGAGUUGGCCUAAUUGGCUGGCCAAGUGUUGGUUAUUUUGGA .......((..(((..((.(((((.((..(((.....)))..)))))))))..)))....)).. ( -24.30, z-score = -3.67, R) >droSec1.super_0 3193102 64 + 21120651 GAGUCCACAUGAACUGACCACUUGAGUUGGCCUAAUUGGCUGGCCAAGUGUUGGUUAUUUUGGA ...((((....(((..((.(((((.((..(((.....)))..)))))))))..)))....)))) ( -27.90, z-score = -4.60, R) >droSim1.chr3R 10156986 64 + 27517382 GAGUCCACAUGAACUGACCACUUGAGUUGGCCUAAUUGGCUGGCCAAGUGUUGGUUAUUUUGGA ...((((....(((..((.(((((.((..(((.....)))..)))))))))..)))....)))) ( -27.90, z-score = -4.60, R) >droVir3.scaffold_13047 9912615 58 - 19223366 GAAAGCACACGUUCUGGCCACUAGUGUUGGCCUAAUUGGCUGGCCAAGUGAAGGAUGU------ ........(((((((...((((.(.((..(((.....)))..))).)))).)))))))------ ( -21.30, z-score = -2.11, R) >droMoj3.scaffold_6540 24685519 58 + 34148556 CAAAGCACACGUUCUGCCCACUUGUGUUGGCCUAAUUGGCUGGCCAAGUGAAGGAUGU------ ........(((((((...((((((.((..(((.....)))..)))))))).)))))))------ ( -25.40, z-score = -3.57, R) >droGri2.scaffold_14906 6639300 58 + 14172833 UCAAGCACACGUUCUGCCCACUUGUGUUGGCCUAAUUGGCUGGCCAAGUGAAGGAUGU------ ........(((((((...((((((.((..(((.....)))..)))))))).)))))))------ ( -25.40, z-score = -3.49, R) >consensus GAGUCCACAUGAACUGACCACUUGAGUUGGCCUAAUUGGCUGGCCAAGUGUUGGUUAUUUUGGA ............(((...((((((.((((.((.....)).))))))))))..)))......... (-11.68 = -11.27 + -0.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:57:20 2011