| Sequence ID | dm3.chr3R |
|---|---|
| Location | 3,837,979 – 3,838,097 |
| Length | 118 |
| Max. P | 0.934793 |

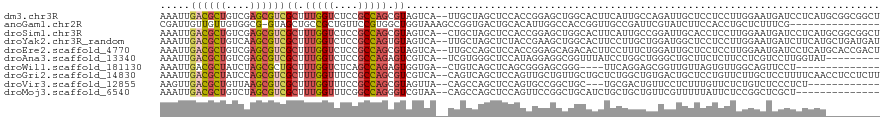
| Location | 3,837,979 – 3,838,097 |
|---|---|
| Length | 118 |
| Sequences | 10 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 59.16 |
| Shannon entropy | 0.87862 |
| G+C content | 0.55430 |
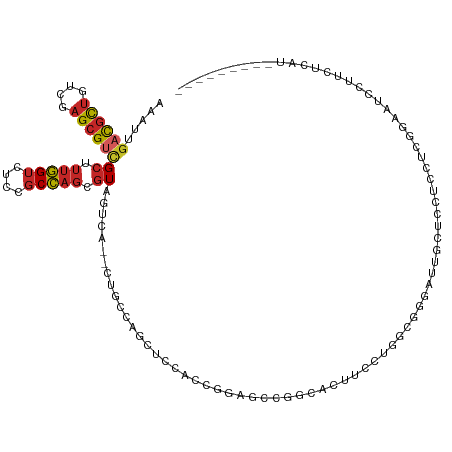
| Mean single sequence MFE | -38.26 |
| Consensus MFE | -10.58 |
| Energy contribution | -11.04 |
| Covariance contribution | 0.46 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.39 |
| SVM RNA-class probability | 0.934793 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 3837979 118 + 27905053 AAAUUGACGCUGUCGAGCGUCGCUUUGGUCUCCGCCAGCGUAGUCA--UUGCUAGCUCCACCGGAGCUGGCACUUCAUUGCCAGAUUGCUCCUCCUUGGAAUGAUCCUCAUGCGGCGGCU .....((((((....))))))..........(((((.(((((((..--.((((((((((...))))))))))....)))))..((((..(((.....)))..)))).....))))))).. ( -46.80, z-score = -2.51, R) >anoGam1.chr2R 45018173 104 - 62725911 CGAUUGUUGUUGUGGCG-GUAGCUGCCGCUGUUCCGUGGCUGGUAAAGCCGGUGACUGCACAUUGGCCACCGGUUGCCGAUUCGUAUCUUCCACCUGCUCUUUCG--------------- (((..(..((.((((.(-(((((((((((......))))).(((...(((((((......))))))).))))))))))(((....)))..))))..)).)..)))--------------- ( -36.90, z-score = -1.33, R) >droSim1.chr3R 3880587 118 + 27517382 AAAUUGACGCUGUCGAGCGUCGCUUUGGUCUCCGCCAGCGUAGUCA--CUGCUAGCUCCACCGGAGCUGGCACUUCAUUGCCGGAUUGCACCUCCUUGGAAUGAUCCUCAUGCGGCGGCU .....((((((....))))))..........(((((.(((((((..--.((((((((((...))))))))))....))))).((((((..((.....))..))))))....))))))).. ( -47.10, z-score = -2.20, R) >droYak2.chr3R_random 1244995 118 + 3277457 AAAUUGACGCUGUCAAGCGUCGCUUUGGUCUCCGCCAGUGUAGUCA--UUGCUAGCUCUACCGAAGCUGGCACUUCCUUGCUGGAUGGCUCCUCCUUGGAAUGAUCUUCAUGCUGAUGAU .....((((((....))))))((...((((.(((((((((.((..(--.(((((((((....).)))))))).)..)))))))).))).(((.....)))..)))).....))....... ( -38.60, z-score = -1.92, R) >droEre2.scaffold_4770 4078290 118 + 17746568 AAAUUGACGCUGUCGAGCGUCGCUUUGGUCUCCGCCAGCGUAGUCA--UUGCCAGCUCCACCGGAGCAGACACUUCCUUUCUGGAUUGCUCCUCCUUGGAAUGAUCCUCAUGCACCGACU .....((((((....))))))((.(((((....))))).))((((.--.(((..(((((...)))))...............(((((..(((.....)))..)))))....)))..)))) ( -37.90, z-score = -1.80, R) >droAna3.scaffold_13340 10546322 109 + 23697760 AAAUUGACGCUGUCGAGCGUCGCUUUGGUCUCCGCCAGAGUCGUCA--UCGUGGGCUCCAUAGGAGGCGGUUUAUCCUGGCUGGGCUGCUUCUCUUCCUCGUCCUUGGUAU--------- .....((((((....))))))((((((((....))))))))....(--(((.((((.....(((((((((((((.......))))))))))).)).....)))).))))..--------- ( -39.10, z-score = -1.23, R) >droWil1.scaffold_181130 8579693 100 - 16660200 AAAUUGACGCUAUCUAGCGCUGCUUUGGUCUCAGCCAGAGUGGUGA--CUGUCAGCUCAGCGGGAGCGGG----UUCAGGAGCGGUUGUUAGUGUUGGCAGUUCCU-------------- ........((((.(((((((..(((((((....)))))))..))((--(((((.((((.((....)))))----)....)).))))))))))...)))).......-------------- ( -35.50, z-score = -0.48, R) >droGri2.scaffold_14830 2500532 118 + 6267026 AAAUUGACGCUAUCCAGCGUCGCUUUGGUUUCCGCCAGCGUCGUCA--CAGUCAGCUCCAGUUGCUGUUGCUGCUCUGGCUGUGACUGCUCCUGUUCUUGCUCCUUUUCAACCUCCUCUU .....((((((....))))))((.(((((....))))).((.((((--(((((((...((((.......))))..))))))))))).))..........))................... ( -37.10, z-score = -3.43, R) >droVir3.scaffold_12855 8861791 103 + 10161210 AAGUUGACGCUGUUAAGCGUCGCUUUGGUUUCCGCCAGCGUAGUUA--CAGCCAGCUCCAGUGCCGGCUGC---UGCGACUGUUCCUCUUUGUUCUCUGUCUCCCUCU------------ .((((((((((....))))))(((..(((....))))))((((...--(((((.((......)).))))))---)))))))...........................------------ ( -31.00, z-score = -1.49, R) >droMoj3.scaffold_6540 24020187 104 + 34148556 AAAUUGACGCUGUCUAGCGUCGCUUUGGUUUCGGCCAGGGUCGUAA--CAGCCAGCUCCAGUUCCGGCUGCAUCUGCUGCUGUUCGUUUUUAUUCUCCGGCUCGCU-------------- .....((((((....))))))((..((((....))))((((((.((--((((.(((..((((....)))).....))))))))).(........)..)))))))).-------------- ( -32.60, z-score = -0.93, R) >consensus AAAUUGACGCUGUCGAGCGUCGCUUUGGUCUCCGCCAGCGUAGUCA__CUGCCAGCUCCACCGGAGCCGGCACUUCCUGGCGGGAUUGCUCCUCCUCGGAAUCCUUCUCAU_________ .....((((((....))))))((.(((((....))))).))............................................................................... (-10.58 = -11.04 + 0.46)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:57:15 2011