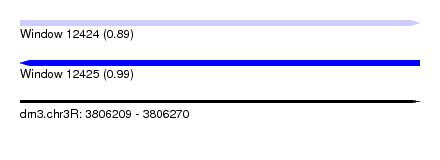
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 3,806,209 – 3,806,270 |
| Length | 61 |
| Max. P | 0.989902 |

| Location | 3,806,209 – 3,806,270 |
|---|---|
| Length | 61 |
| Sequences | 4 |
| Columns | 63 |
| Reading direction | forward |
| Mean pairwise identity | 87.80 |
| Shannon entropy | 0.19316 |
| G+C content | 0.39970 |
| Mean single sequence MFE | -16.05 |
| Consensus MFE | -13.50 |
| Energy contribution | -13.50 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.08 |
| SVM RNA-class probability | 0.887581 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
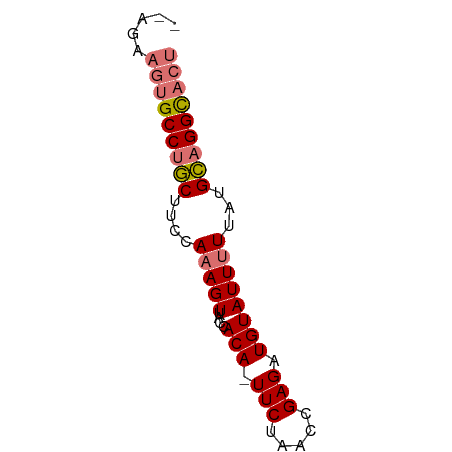
>dm3.chr3R 3806209 61 + 27905053 AGUGCCUGCAUAAGAAUACAUCUCGGUUAGAAAUGUGGUAACUUUGGAAGCAGGCACUUCU-- (((((((((..(((..(((((.((.....)).)))))....))).....)))))))))...-- ( -20.70, z-score = -3.18, R) >droYak2.chr3R 22365399 62 - 28832112 AGUACCGUCAUAAAAAUACAUCUCGGUUAGAA-UGUGAUAACUUUGGAAGAAGGCAUAUUAUU ...((((..((........))..))))...((-((((....((((....)))).))))))... ( -7.90, z-score = 0.08, R) >droSec1.super_6 3874595 60 + 4358794 AGAGCCUGCAUAAAAAUACAUCUCGGUUAGAA-UGUGGUAACUUUGGAGGCAGGCACUUCU-- ...((((((.((((..(((((.((.....)))-)))).....))))...))))))......-- ( -16.30, z-score = -1.64, R) >droSim1.chr3R 3848818 60 + 27517382 AGUGCCUGCAUAAAAAUACAUCUCGGUUAGAA-UGUGGUAACUUUGGAAGCAGGCACUUCU-- (((((((((.((((..(((((.((.....)))-)))).....))))...)))))))))...-- ( -19.30, z-score = -3.03, R) >consensus AGUGCCUGCAUAAAAAUACAUCUCGGUUAGAA_UGUGGUAACUUUGGAAGCAGGCACUUCU__ .((((((((.((((..((((..((.....))..)))).....))))...))))))))...... (-13.50 = -13.50 + 0.00)

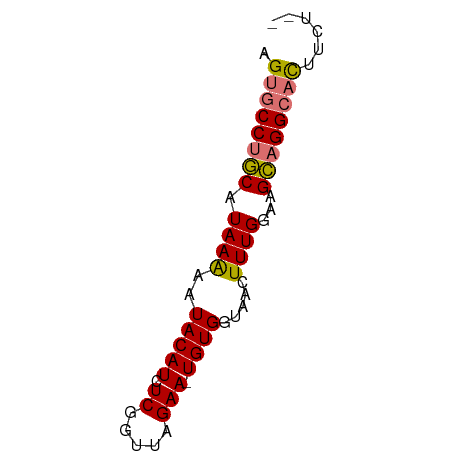
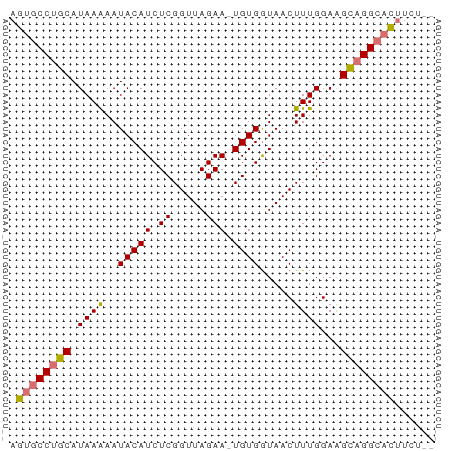
| Location | 3,806,209 – 3,806,270 |
|---|---|
| Length | 61 |
| Sequences | 4 |
| Columns | 63 |
| Reading direction | reverse |
| Mean pairwise identity | 87.80 |
| Shannon entropy | 0.19316 |
| G+C content | 0.39970 |
| Mean single sequence MFE | -15.03 |
| Consensus MFE | -13.06 |
| Energy contribution | -13.75 |
| Covariance contribution | 0.69 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.989902 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
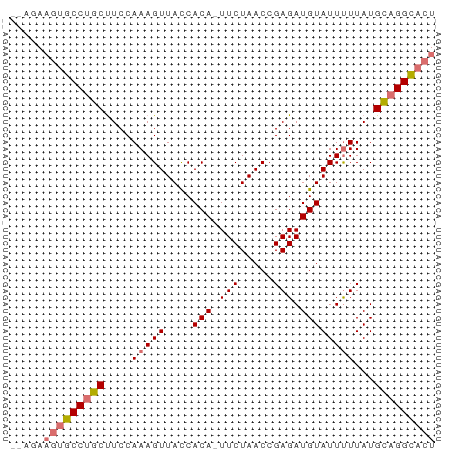
>dm3.chr3R 3806209 61 - 27905053 --AGAAGUGCCUGCUUCCAAAGUUACCACAUUUCUAACCGAGAUGUAUUCUUAUGCAGGCACU --...(((((((((.....(((.....(((((((.....)))))))...)))..))))))))) ( -22.00, z-score = -5.10, R) >droYak2.chr3R 22365399 62 + 28832112 AAUAAUAUGCCUUCUUCCAAAGUUAUCACA-UUCUAACCGAGAUGUAUUUUUAUGACGGUACU .......((((.((....(((((((((...-..........)))).)))))...)).)))).. ( -6.22, z-score = -0.06, R) >droSec1.super_6 3874595 60 - 4358794 --AGAAGUGCCUGCCUCCAAAGUUACCACA-UUCUAACCGAGAUGUAUUUUUAUGCAGGCUCU --...((.((((((....(((((....(((-((........))))))))))...)))))).)) ( -13.60, z-score = -1.84, R) >droSim1.chr3R 3848818 60 - 27517382 --AGAAGUGCCUGCUUCCAAAGUUACCACA-UUCUAACCGAGAUGUAUUUUUAUGCAGGCACU --...(((((((((....(((((....(((-((........))))))))))...))))))))) ( -18.30, z-score = -3.82, R) >consensus __AGAAGUGCCUGCUUCCAAAGUUACCACA_UUCUAACCGAGAUGUAUUUUUAUGCAGGCACU .....(((((((((....(((((....(((.(((.....))).))))))))...))))))))) (-13.06 = -13.75 + 0.69)
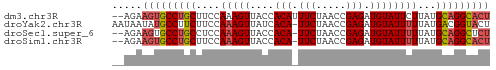
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:57:12 2011