
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 3,761,274 – 3,761,385 |
| Length | 111 |
| Max. P | 0.920706 |

| Location | 3,761,274 – 3,761,385 |
|---|---|
| Length | 111 |
| Sequences | 14 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.90 |
| Shannon entropy | 0.35905 |
| G+C content | 0.43384 |
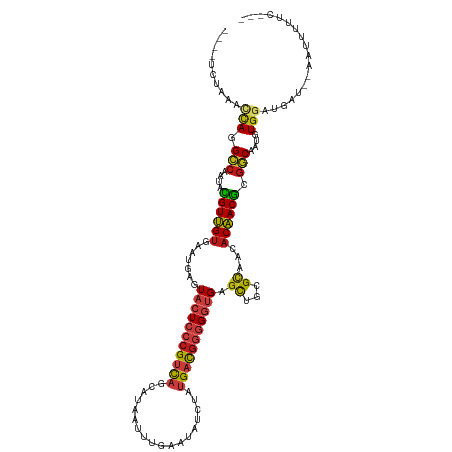
| Mean single sequence MFE | -32.02 |
| Consensus MFE | -21.53 |
| Energy contribution | -21.88 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.920706 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
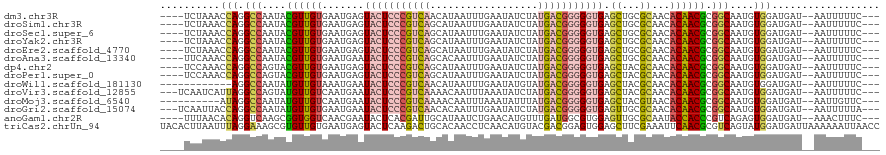
>dm3.chr3R 3761274 111 - 27905053 ----UCUAAACCAGGCCAAUACGUUGUGAAUGAGUACUCCCGUCAACAUAAUUUGAAUAUCUAUGACGGGGGUGAGCUGCGCAACACAACGCGGCAAUGUGGAUGAU--AAUUUUUC--- ----......(((.(((....(((((((......(((((((((((..................))))))))))).((...))..))))))).)))....))).....--........--- ( -32.17, z-score = -1.97, R) >droSim1.chr3R 3799097 111 - 27517382 ----UCUAAACCAGGCCAAUACGUUGUGAAUGAGUACUCCCGUCAGCAUAAUUUGAAUAUCUAUGACGGGGGUGAGCUGCGCAACACAACGCGGCAAUGUGGAUGAU--AAUUUUUC--- ----......(((.(((....(((((((......(((((((((((..(((.......)))...))))))))))).((...))..))))))).)))....))).....--........--- ( -33.20, z-score = -1.95, R) >droSec1.super_6 3831222 111 - 4358794 ----UCUAAACCAGGCCAAUACGUUGUGAAUGAGUACUCCCGUCAGCAUAAUUUGAAUAUCUAUGACGGGGGUGAGCUGCGCAACACAACGCGGCAAUGUGGAUGAU--AAUUUUUC--- ----......(((.(((....(((((((......(((((((((((..(((.......)))...))))))))))).((...))..))))))).)))....))).....--........--- ( -33.20, z-score = -1.95, R) >droYak2.chr3R 22320080 111 + 28832112 ----UCUAAACCAGGCCAAUACGUUGUGAAUGAGUACUCCCGUCAGCAUAAUUUGAAUAUCUAUGACGGGGGUGAGCUGCGCAACACAACGCGGCAAUGUGGAUGAU--AAUUUUUC--- ----......(((.(((....(((((((......(((((((((((..(((.......)))...))))))))))).((...))..))))))).)))....))).....--........--- ( -33.20, z-score = -1.95, R) >droEre2.scaffold_4770 4001686 111 - 17746568 ----UCUAAACCAGGCCAAUACGUUGUGAAUGAGUACUCCCGUCAGCAUAAUUUGAAUAUCUAUGACGGGGGUGAGCUGCGCAACACAACGCGGCAAUGUGGAUGAU--AAUUUUUC--- ----......(((.(((....(((((((......(((((((((((..(((.......)))...))))))))))).((...))..))))))).)))....))).....--........--- ( -33.20, z-score = -1.95, R) >droAna3.scaffold_13340 14871925 111 - 23697760 ----UUCAAACCAGGCCAAUACGUUGUGAAUGAAUACUCCCGUCAGCACAAUUUGAAUAUCUAUGACGGGGGUGAGCUGCGCAACACAACGCGGCAAUGUGGAUGAU--AAUUUUUC--- ----......(((.(((....(((((((......(((((((((((..................))))))))))).((...))..))))))).)))....))).....--........--- ( -33.07, z-score = -2.04, R) >dp4.chr2 13768732 111 + 30794189 ----UCCAAACCAGGCCAGUACGUUGUGAAUGAGUACUCCCGUCAGCAUAAUUUGAAUAUCUAUGACGGGGGUGAGCUACGCAACACAACGCGGCAAUGUGGAUGAU--AAUUUUUC--- ----((((......(((.((..((((((...(..(((((((((((..(((.......)))...)))))))))))..)..))))))...))..)))....))))....--........--- ( -35.90, z-score = -2.66, R) >droPer1.super_0 3363936 111 - 11822988 ----UCCAAACCAGGCCAGUACGUUGUGAAUGAGUACUCCCGUCAGCAUAAUUUGAAUAUCUAUGACGGGGGUGAGCUACGCAACACAACGCGGCAAUGUGGAUGAU--AAUUUUUC--- ----((((......(((.((..((((((...(..(((((((((((..(((.......)))...)))))))))))..)..))))))...))..)))....))))....--........--- ( -35.90, z-score = -2.66, R) >droWil1.scaffold_181130 3461780 103 + 16660200 ------------AGGCCAAUAUGUUGUAAAUGAAUACUCCCGUCAACAUAAUUUGAAUAUGUAUGACGGGGGUGAGCUACGCAACACAACGCGGCAAUGUGGAUGAU--AAUUUUUC--- ------------..(((....((((((....(..((((((((((((((((.......))))).)))))))))))..)...))))))......)))............--........--- ( -31.30, z-score = -2.84, R) >droVir3.scaffold_12855 2028367 112 + 10161210 ---UCAAUCAUUAGGCCAGUAUGUUGUCAAUGAAUACUCCCGUCAAAACAAUUUAAAUAUCUAUGACGGGGGUGAGCUACGCAACACAACGCGGCAAUGUGGAUGAU--AAUUUUUC--- ---(((.((((...(((.((.((((((....(..(((((((((((..................)))))))))))..)...))))))..))..)))...)))).))).--........--- ( -33.77, z-score = -3.47, R) >droMoj3.scaffold_6540 10781941 105 + 34148556 ----------AUAGGCCAAUAUGUUGUCAAUGAAUACUCCCGUCAAAACAAUUUAAAUAUUUAUGACGGGGGUGAGCUACGUAACACAACGCGGCAAUGUGGAUGAU--AAUUGUUC--- ----------...(..((((.(((..((......(((((((((((..................))))))))))).((..(((......)))..))......))..))--)))))..)--- ( -30.17, z-score = -2.67, R) >droGri2.scaffold_15074 3003794 112 + 7742996 ---UCAAUUACCAGGCCAAUAUGUUGUGAAUGAAUACUCCCGUCAACACAAUUUGAAUAUCUAUGACGGGGGUGAGUUGCGCAACACAACGCGGCAAUGUGGAUGAU--AAUUUUUA--- ---(((.((((...(((....((((((((((...(((((((((((..................))))))))))).))).)))))))......)))...)))).))).--........--- ( -32.97, z-score = -2.19, R) >anoGam1.chr2R 50510363 111 - 62725911 ----UUUAACACAGGUCAAGCGGUGGUCAACGAAUACUCACGAUUGCAUAAUCUGAACAUGUUUGAUGGCGUGGAGUUGCGCAAUACCACCCGUCAGAGUGGAUGAU--AAACUUUC--- ----.......(((((...(((((.((............)).)))))...)))))..(((.((((((((.((((.(((....))).)))))))))))))))......--........--- ( -27.80, z-score = -0.20, R) >triCas2.chrUn_94 167671 120 + 392035 UACACUUAAUUUAGGAAAGCGUGUUGUGAAUGAGUACUCAAGACUGCACAACCUCAACAUGUACGACGGAGUGGAGCUUCGAAAUUCAACGCGUCAGUAUGGAUGAUUAAAAAAUUAACC .....(((((((..(((.((((((((.(..((.(((........))).))..).))))))))....(((((.....)))))...)))....((((......))))......))))))).. ( -22.40, z-score = 0.61, R) >consensus ____UCUAAACCAGGCCAAUACGUUGUGAAUGAGUACUCCCGUCAGCAUAAUUUGAAUAUCUAUGACGGGGGUGAGCUGCGCAACACAACGCGGCAAUGUGGAUGAU__AAUUUUUC___ ..........(((.(((....((((((.......(((((((((((..................))))))))))).((...))...)))))).)))....))).................. (-21.53 = -21.88 + 0.35)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:57:07 2011