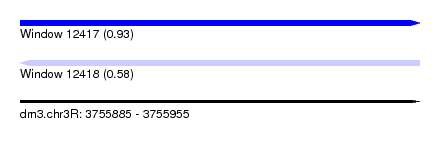
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 3,755,885 – 3,755,955 |
| Length | 70 |
| Max. P | 0.931051 |

| Location | 3,755,885 – 3,755,955 |
|---|---|
| Length | 70 |
| Sequences | 9 |
| Columns | 73 |
| Reading direction | forward |
| Mean pairwise identity | 87.89 |
| Shannon entropy | 0.24403 |
| G+C content | 0.58273 |
| Mean single sequence MFE | -26.27 |
| Consensus MFE | -20.79 |
| Energy contribution | -20.54 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.931051 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 3755885 70 + 27905053 UACUUACACGAAGAGGACAGCGGCCGCUGCUGCUGCUGCCACGGCGGCAGCAACGUUGCGAUACUGCUAC--- ..................(((((.(((.(((((((((((....)))))))))..)).)))...)))))..--- ( -28.00, z-score = -2.10, R) >droGri2.scaffold_15074 2997037 70 - 7742996 UACUUACACAAAGAGUACAGCUGCAGC---AGCUGCCGCAGCAGCGGCGGCAACAUUGCGAUACUGCUGCUAC (((((.......))))).....(((((---((.((((((....))))))(((....)))....)))))))... ( -26.30, z-score = -1.66, R) >droWil1.scaffold_181130 3453890 66 - 16660200 -ACUUACACAAAGAGCACAGCUGCCGCUGCAGCUGCUGCCACAGCGGCAGCAACAUUUCUAUACUAC------ -..........((((....((((((((((..((....))..)))))))))).....)))).......------ ( -25.10, z-score = -3.76, R) >droPer1.super_0 3358051 70 + 11822988 UACUUACACAAAGAGGACAGCUGCCGCUGCUGCUGCCGCCACAGCGGCGGCAACAUUGCGAUACUGCUAC--- ..............((.(((.((.(((((.(((((((((....))))))))).))..))).)))))))..--- ( -25.10, z-score = -2.04, R) >dp4.chr2 13763075 70 - 30794189 UACUUACACAAAGAGGACAGCUGCCGCUGCUGCUGCCGCCACAGCGGCGGCAACAUUGCGAUACUGCUAC--- ..............((.(((.((.(((((.(((((((((....))))))))).))..))).)))))))..--- ( -25.10, z-score = -2.04, R) >droAna3.scaffold_13340 14866850 70 + 23697760 UACUUACACAAAAAGGACAGCUGCCGCUGCUGCUGCUGCCACGGCGGCAGCAACGUUGCGAUAUUGCUAC--- ...................((((((((((..((....))..))))))))))...((.(((....))).))--- ( -25.00, z-score = -1.79, R) >droEre2.scaffold_4770 3996196 70 + 17746568 UACUUACACGAAGAGGACAGCGGCCGCUGCUGCUGCUGCCACGGCGGCAGCAACGUUGCGAUACUGCUAC--- ..................(((((.(((.(((((((((((....)))))))))..)).)))...)))))..--- ( -28.00, z-score = -2.10, R) >droSec1.super_6 3825801 70 + 4358794 UACUUACACGAAGAGGACAGCGGCUGCUGCUGCUGCUGCCACGGCGGCAGCAACGUUGCGAUACUGCUAC--- ...........((((....((((((((((((((((......)))))))))))..)))))....)).))..--- ( -26.90, z-score = -1.84, R) >droSim1.chr3R 3793716 70 + 27517382 UACUUACACGAAGAGGACAGCGGCUGCUGCUGCUGCUGCCACGGCGGCAGCAACGUUGCGAUACUGCUAC--- ...........((((....((((((((((((((((......)))))))))))..)))))....)).))..--- ( -26.90, z-score = -1.84, R) >consensus UACUUACACAAAGAGGACAGCUGCCGCUGCUGCUGCUGCCACGGCGGCAGCAACGUUGCGAUACUGCUAC___ ......(.(((......((((....))))..((((((((....))))))))....))).)............. (-20.79 = -20.54 + -0.25)

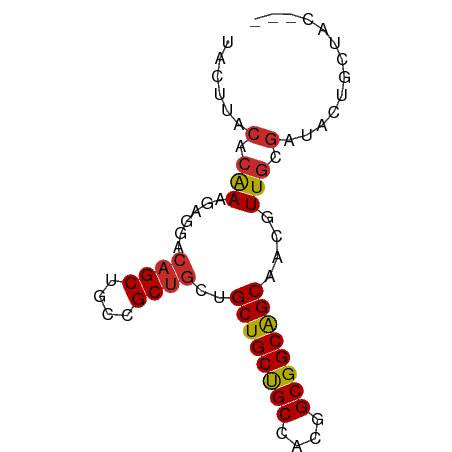
| Location | 3,755,885 – 3,755,955 |
|---|---|
| Length | 70 |
| Sequences | 9 |
| Columns | 73 |
| Reading direction | reverse |
| Mean pairwise identity | 87.89 |
| Shannon entropy | 0.24403 |
| G+C content | 0.58273 |
| Mean single sequence MFE | -28.02 |
| Consensus MFE | -23.18 |
| Energy contribution | -23.40 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.584920 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 3755885 70 - 27905053 ---GUAGCAGUAUCGCAACGUUGCUGCCGCCGUGGCAGCAGCAGCAGCGGCCGCUGUCCUCUUCGUGUAAGUA ---...(((((.((((...((((((((.((....)).)))))))).))))..)))))................ ( -27.00, z-score = -0.86, R) >droGri2.scaffold_15074 2997037 70 + 7742996 GUAGCAGCAGUAUCGCAAUGUUGCCGCCGCUGCUGCGGCAGCU---GCUGCAGCUGUACUCUUUGUGUAAGUA ((((((((......(((....))).(((((....))))).)))---))))).(((.(((.......)))))). ( -29.60, z-score = -1.09, R) >droWil1.scaffold_181130 3453890 66 + 16660200 ------GUAGUAUAGAAAUGUUGCUGCCGCUGUGGCAGCAGCUGCAGCGGCAGCUGUGCUCUUUGUGUAAGU- ------....((((.(((.((.((((((((((..((....))..))))))))))...))..))).))))...- ( -32.40, z-score = -3.33, R) >droPer1.super_0 3358051 70 - 11822988 ---GUAGCAGUAUCGCAAUGUUGCCGCCGCUGUGGCGGCAGCAGCAGCGGCAGCUGUCCUCUUUGUGUAAGUA ---...(((((.((((..((((((((((.....))))))))))...))))..)))))................ ( -29.50, z-score = -1.44, R) >dp4.chr2 13763075 70 + 30794189 ---GUAGCAGUAUCGCAAUGUUGCCGCCGCUGUGGCGGCAGCAGCAGCGGCAGCUGUCCUCUUUGUGUAAGUA ---...(((((.((((..((((((((((.....))))))))))...))))..)))))................ ( -29.50, z-score = -1.44, R) >droAna3.scaffold_13340 14866850 70 - 23697760 ---GUAGCAAUAUCGCAACGUUGCUGCCGCCGUGGCAGCAGCAGCAGCGGCAGCUGUCCUUUUUGUGUAAGUA ---..........(((((.(((((((((.....))))))))).(((((....))))).....)))))...... ( -26.80, z-score = -1.11, R) >droEre2.scaffold_4770 3996196 70 - 17746568 ---GUAGCAGUAUCGCAACGUUGCUGCCGCCGUGGCAGCAGCAGCAGCGGCCGCUGUCCUCUUCGUGUAAGUA ---...(((((.((((...((((((((.((....)).)))))))).))))..)))))................ ( -27.00, z-score = -0.86, R) >droSec1.super_6 3825801 70 - 4358794 ---GUAGCAGUAUCGCAACGUUGCUGCCGCCGUGGCAGCAGCAGCAGCAGCCGCUGUCCUCUUCGUGUAAGUA ---((.((......)).))(((((((((.....))))))))).(((((....)))))................ ( -25.20, z-score = -0.77, R) >droSim1.chr3R 3793716 70 - 27517382 ---GUAGCAGUAUCGCAACGUUGCUGCCGCCGUGGCAGCAGCAGCAGCAGCCGCUGUCCUCUUCGUGUAAGUA ---((.((......)).))(((((((((.....))))))))).(((((....)))))................ ( -25.20, z-score = -0.77, R) >consensus ___GUAGCAGUAUCGCAACGUUGCUGCCGCCGUGGCAGCAGCAGCAGCGGCAGCUGUCCUCUUUGUGUAAGUA .............(((...(((((((((.....))))))))).(((((....))))).......)))...... (-23.18 = -23.40 + 0.22)
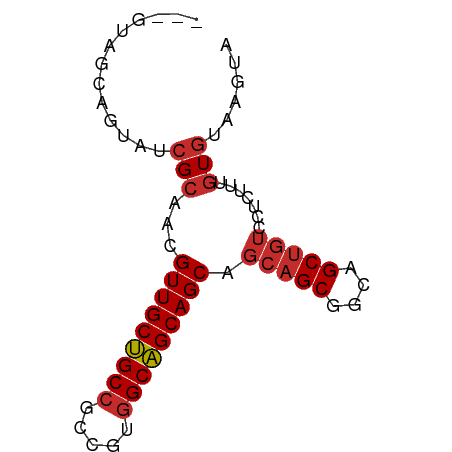
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:57:06 2011