| Sequence ID | dm3.chr3R |
|---|---|
| Location | 3,707,091 – 3,707,194 |
| Length | 103 |
| Max. P | 0.565979 |

| Location | 3,707,091 – 3,707,194 |
|---|---|
| Length | 103 |
| Sequences | 9 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 67.82 |
| Shannon entropy | 0.67556 |
| G+C content | 0.50990 |
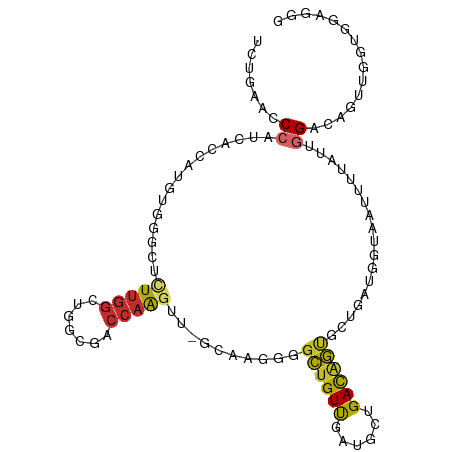
| Mean single sequence MFE | -34.56 |
| Consensus MFE | -7.78 |
| Energy contribution | -7.22 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.69 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.23 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.565979 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
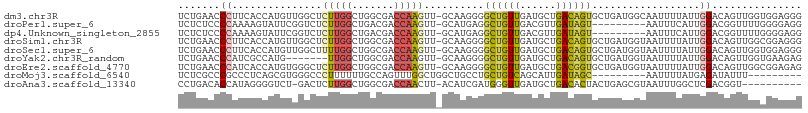
>dm3.chr3R 3707091 103 - 27905053 UCUGAACCCUUCACCAUGUUGGCUCUUGGCUGGCGACCAAGUU-GCAAGGGGCUGUUGAUGCUGACAGUGCUGAUGGCAAUUUUAUUGGACAGUUGGUGGAGGG ......(((((((((((((..((.....))..))).(((((((-((..((.((((((......)))))).))....)))))....)))).....)))))))))) ( -43.20, z-score = -3.29, R) >droPer1.super_6 2404895 94 - 6141320 UCUCUCCCCAAAAGUAUUCGGUCUCUUGGCUGACGACCAAGUU-GCAUGAGGCUGUUGACGUUGAUAGU---------AAUUUCAUUGGACGGUUUUGGGGAGG ...((((((((((((...(((((((...((.(((......)))-))..)))))))...))..(((....---------....)))........)))))))))). ( -32.80, z-score = -2.42, R) >dp4.Unknown_singleton_2855 508 94 + 992 UCUCUCCCCAAAAGUAUUCGGUCUCUUGGCUGACGACCAAGUU-GCAUGAGGCUGUUGACGUUGAUAGU---------AAUUUCAUUGGACGGUUUUGGGGAGG ...((((((((((((...(((((((...((.(((......)))-))..)))))))...))..(((....---------....)))........)))))))))). ( -32.80, z-score = -2.42, R) >droSim1.chr3R 3743699 103 - 27517382 UCUGAACCCUUCACCAUGUUGGCUCUUGGCUGGCGACCAAGUU-GCAAGGGGCUGUUGAUGCUGACAGUGCUGAUGGUAAUUUUAUUGGACAGUUGGCGGAGGG ......((((((.((((((..((.....))..))).(((((((-((..((.((((((......)))))).))....)))))....)))).....))).)))))) ( -37.30, z-score = -1.70, R) >droSec1.super_6 3777040 103 - 4358794 UCUGAACCCUUCACCAUGUUGGCUUUUGGCUGGCGACCAAGUU-GCAAGGGGCUGUUGAUGCUGACAGUGCUGAUGGUAAUUUUAUUGGACAGUUGGUGGAGGG ......(((((((((((((..((.....))..))).(((((((-((..((.((((((......)))))).))....)))))....)))).....)))))))))) ( -41.20, z-score = -3.34, R) >droYak2.chr3R_random 1186448 96 - 3277457 UCUGAACCCAUCGCCAUG-------UUGGCUGGCGACCAAGUU-GCAAGGGGCUGUUGAUGCUGACAGUGCUGAUGGUAAUUUUAUUGGACAGUUGGUGAAGAG (((..(((..((((((.(-------....)))))))(((((((-((..((.((((((......)))))).))....)))))....))))......)))..))). ( -30.80, z-score = -1.19, R) >droEre2.scaffold_4770 3948253 103 - 17746568 UCUGAACCCAUCACCAUGUGGGCUCUUGGCUGGCGACCAAGUU-GCAAGGGGCUGUUGAUGCUGACGGUGCUGAUGGUAAUUUUAUUGGACAGUUGGCGGAGAG ......(((((......)))))(((((.((..((..(((((((-((..((.((((((......)))))).))....)))))....))))...))..)).))))) ( -36.80, z-score = -1.54, R) >droMoj3.scaffold_6540 7019147 86 - 34148556 UCUCGCCCGCCCUCAGCGUGGGCCCUUUUUUGCCAGUUUGGCUGGCUGCCUGCUGUCAGCAUUGAUAGC---------AAUUUUAUGAGAUAUUU--------- ((((((((((.......))))))........(((((.....)))))....(((((((......))))))---------).......)))).....--------- ( -31.60, z-score = -2.57, R) >droAna3.scaffold_13340 10890041 92 - 23697760 CCUGACACCAUAGGGGUCU-GACUCUUGGCUGGCGACCAACUU-ACAUCGAUGGGUUGAUGCUGACACUACUGAGCGUAAUUUGGCUCGACGGU---------- .(((.(((((.((((....-..))))))).)).(((((((.((-((.(((.((((((......))).))).)))..)))).)))).))).))).---------- ( -24.50, z-score = -0.27, R) >consensus UCUGAACCCAUCACCAUGUGGGCUCUUGGCUGGCGACCAAGUU_GCAAGGGGCUGUUGAUGCUGACAGUGCUGAUGGUAAUUUUAUUGGACAGUUGGUGGAGGG ...........................(((((....(((((....).....((((((......))))))................)))).)))))......... ( -7.78 = -7.22 + -0.56)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:57:01 2011