| Sequence ID | dm3.chr3R |
|---|---|
| Location | 3,698,886 – 3,699,004 |
| Length | 118 |
| Max. P | 0.521298 |

| Location | 3,698,886 – 3,699,004 |
|---|---|
| Length | 118 |
| Sequences | 8 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 74.06 |
| Shannon entropy | 0.49073 |
| G+C content | 0.47358 |
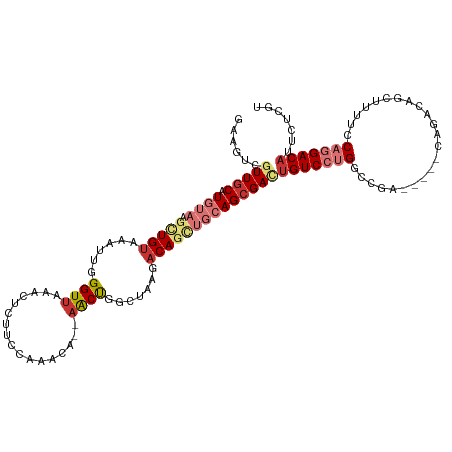
| Mean single sequence MFE | -31.11 |
| Consensus MFE | -15.85 |
| Energy contribution | -17.95 |
| Covariance contribution | 2.10 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.29 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.521298 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
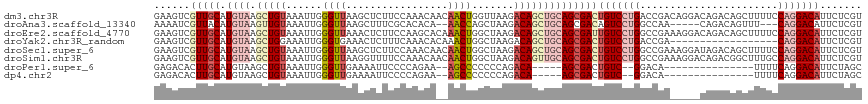
>dm3.chr3R 3698886 118 + 27905053 GAAGUCGUUGCAUGUAAGCUGUAAAUUGGGUUAAGCUCUUCCAAACAACAACUGGUUAAGACAGCUGCAGCGACUGUCCUGACCGACAGGACAGACAGCUUUUCCAGGACAUUCUCGU (((((((((((.((((.(((((...(((((.........))))).......((.....))))))))))))))))(((((((.....)))))))..............))).))).... ( -35.00, z-score = -1.26, R) >droAna3.scaffold_13340 10884460 108 + 23697760 AAAAUCGUUACAUGUAAGUUGUAAAUUGGGUUAAGCUUUCGCACACA--AACCAGCUAAGACAGCUGCAGCGACAGUCCUGGCCAA-----CAGACAGUUU---CAGGACAUUCUCGU ......((..(.((((.(((((......((((..((....)).....--)))).......))))))))))..)).(((((((..((-----(.....))))---))))))........ ( -24.82, z-score = -0.45, R) >droEre2.scaffold_4770 3938540 118 + 17746568 GAAGUCGUUGCAUGUAAGCUGUAAAUUGGGUUAAACUCUUCCAAGCACAAACUGGCUAAGACAGCUGCAGCGAUUGUCCUGGCCGAAAGGACAGACAGCUUUUCCAGGACAUUCUCGU ..((((((((((.....(((((....((((.........))))(((........)))...)))))))))))))))(((((((((....))..((....))...)))))))........ ( -38.10, z-score = -1.98, R) >droYak2.chr3R_random 1182122 100 + 3277457 GAAGUCGUUGCAUGUAAGCUGGAAAUUGGGUGAAACUCUUUCAAACACAAACUGGCUAAGACAGCUGCAGCGACUGUCCUGACCGA------------------CAGGACAUUCUCGU ..(((((((((((((.(((..(....((..((((.....))))..))....)..)))...)))..))))))))))((((((.....------------------))))))........ ( -37.10, z-score = -3.71, R) >droSec1.super_6 3768994 118 + 4358794 GAAGUCGUUGCAUGUAAGCUGUAAAUUGGGUUAAGCUCUUCCAAACAACAACUGGCUAAGACAGCUGCAGCGACUGUCCUGGCCGAAAGGAUAGACAGCUUUUCCAGGACAUUCUCGU ..((((((((((.....(((((...(((((.........))))).......((.....)))))))))))))))))(((((((((....)).............)))))))........ ( -39.01, z-score = -2.27, R) >droSim1.chr3R 3735003 118 + 27517382 GAAGUCGUUGCAUGUAAGCUGUAAAUUGGGUUAAGGUUUUCCAAACAACAACUGGCUAAGACAGUUGCAGCGACUGUCCUGGCCGAAAGGACAGACGGCUUUGCCAGGACAUUCUCGU ..((((((((((.....(((((...((((.(....(((........)))....).)))).)))))))))))))))(((((((((....)).((((....)))))))))))........ ( -40.50, z-score = -2.30, R) >droPer1.super_6 2396157 94 + 6141320 GAGACACUUGCAUGUAAGCUGUAAAUUGGGUUGAAAAUUCCCCAGAA--AGCCCCCCCAGACA-----AGCGACUGUC--GGACA---------------UUUUCAGGACAUUCUAGC ..((((.((((.(((...(((......(((((...............--)))))...))))))-----.)))).))))--(..(.---------------......)..)........ ( -17.16, z-score = 0.82, R) >dp4.chr2 27103690 94 + 30794189 GAGACACUUGCAUGUAAGCUGUAAAUUGGGUUGAAAAUUCCCCAGAA--AGCCCCCCCAGACA-----AGCGACUGUC--GGACA---------------UUUUCAGGACAUUCUAGC ..((((.((((.(((...(((......(((((...............--)))))...))))))-----.)))).))))--(..(.---------------......)..)........ ( -17.16, z-score = 0.82, R) >consensus GAAGUCGUUGCAUGUAAGCUGUAAAUUGGGUUAAACUCUUCCAAACA__AACUGGCUAAGACAGCUGCAGCGACUGUCCUGGCCGA_____CAGACAGCUUUUCCAGGACAUUCUCGU ......(((((.((((.(((((...((((..........................)))).))))))))))))))(((((((.......................)))))))....... (-15.85 = -17.95 + 2.10)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:56:59 2011