
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 3,698,568 – 3,698,666 |
| Length | 98 |
| Max. P | 0.727045 |

| Location | 3,698,568 – 3,698,666 |
|---|---|
| Length | 98 |
| Sequences | 9 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 69.64 |
| Shannon entropy | 0.58639 |
| G+C content | 0.53745 |
| Mean single sequence MFE | -38.88 |
| Consensus MFE | -10.99 |
| Energy contribution | -10.66 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.29 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.727045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 3698568 98 + 27905053 ---------------CUUUUGGGCCCAGGGCUAUUGGCC-AAUUGGCUGGUUACUCAAACAGUUUCGUUUCGGGCUCUCGGCCAAAUUGCGGCUUGC--CGCAUUU-CAAUGCGGCA-- ---------------.....((((((.(((..((..(((-....)))..))..)))((((......)))).))))))..((((.......))))(((--(((((..-..))))))))-- ( -41.00, z-score = -3.06, R) >dp4.chr2 27103384 112 + 30794189 --CCGACCCGAGCCCCUUUUGGCCAGAAGCGAACCGAACCAAUUGGCCAGUUAGUCAAACGGGUUU----CGGGCGCUCGGCCAAACUGCGGUUUGUGUCGCAUUU-CAAUGCAGCAAA --..(((.(((((((..(((((((.((.(((..(((((((..(((((......)))))...)).))----))).))))))))))))..).)))))).)))((((..-..))))...... ( -44.90, z-score = -2.69, R) >droPer1.super_6 2395852 112 + 6141320 --CCGACCCGAGCCCCUUUUGGCCAGAAGCGAACCGAACCAAUUGGCCAGUUAGUCAAACGGGUUU----CGGGCGCUCGGCCAAACUGCGGUUUGUGUCGCAUUU-CAAUGCAGCAAA --..(((.(((((((..(((((((.((.(((..(((((((..(((((......)))))...)).))----))).))))))))))))..).)))))).)))((((..-..))))...... ( -44.90, z-score = -2.69, R) >droSim1.chr3R 3734588 98 + 27517382 ---------------CUUUUGGGCCCAGGGCUAUUGGCC-AAUUGGCUGGUUAGUCAAACAGUUUCGUUUCGGGCUCUCGGCCAAAUUGCGGCUUGC--CGCAUUU-CAAUGCGGCA-- ---------------.....((((((..((((((..(((-....)))..).)))))((((......)))).))))))..((((.......))))(((--(((((..-..))))))))-- ( -41.80, z-score = -3.07, R) >droSec1.super_6 3768675 98 + 4358794 ---------------CUUUUGGGCCCAGGGCUAUUGGCC-AAUUGGCUGGUUAGUCAAACAGUUUCGUUUCGGGCUCUCGGCCAAAUUGCGGCUUGC--CGCAUUU-CAAUGCGGCA-- ---------------.....((((((..((((((..(((-....)))..).)))))((((......)))).))))))..((((.......))))(((--(((((..-..))))))))-- ( -41.80, z-score = -3.07, R) >droYak2.chr3R_random 1181805 98 + 3277457 ---------------CUUUUAGGCCCAGGGCUGUUGGCC-AAUUGGCUGGUGAGUCAAACAGUUUCGUUUCGGGCUCUCGGCCAAAUUGCGGUUUGC--CGCAUUU-CAAUGCGGCA-- ---------------.....(((((((((((.....)))-..((((((((.(((((.(((......)))...))))))))))))).))).)))))((--(((((..-..))))))).-- ( -41.20, z-score = -2.87, R) >droEre2.scaffold_4770 3938226 98 + 17746568 ---------------CUAUUGGGCCCAGGGCUUUUGGCC-AAUUGGCUGGUUAGUCAAACAGUUUCAUUCCGGGCUCUCGGCCAAAUUGCGGCUUGC--CGCAUUU-CAAUGCGGCA-- ---------------.....((((((..(((.....)))-..((((((....)))))).............))))))..((((.......))))(((--(((((..-..))))))))-- ( -37.60, z-score = -1.60, R) >droAna3.scaffold_13340 10884197 79 + 23697760 ------------------------------CUGCAGACCCAAUUGGCCAGUUAUUCAAACAGUUUC-----GGGCUCUCAGCCAAAUUGCGGUUUGU--CGCAUUU-CAAUGCGGCA-- ------------------------------....(((((((((((((..(((.....)))(((...-----..)))....))).))))).)))))((--(((((..-..))))))).-- ( -22.60, z-score = -1.22, R) >droGri2.scaffold_14906 2809059 105 + 14172833 CGGGGAAAAUAGUCAUCGCUGAAGCCAAAGC---UGGCGCAAUUGGCCAGUUUAGAUGGCAAAU------CGGGCGCGUUGCCAAAACGUGG-UUGU--CACAUUUGCAAUGCAGCA-- ...........((((((((....))..((((---((((.......)))))))).))))))....------...(((((((((..(((.(((.-....--))).)))))))))).)).-- ( -34.10, z-score = -0.38, R) >consensus _______________CUUUUGGGCCCAGGGCUAUUGGCC_AAUUGGCUGGUUAGUCAAACAGUUUC_UU_CGGGCUCUCGGCCAAAUUGCGGCUUGC__CGCAUUU_CAAUGCGGCA__ ............................................((((((((.....)))............(((.....)))......)))))(((...((((.....)))).))).. (-10.99 = -10.66 + -0.33)
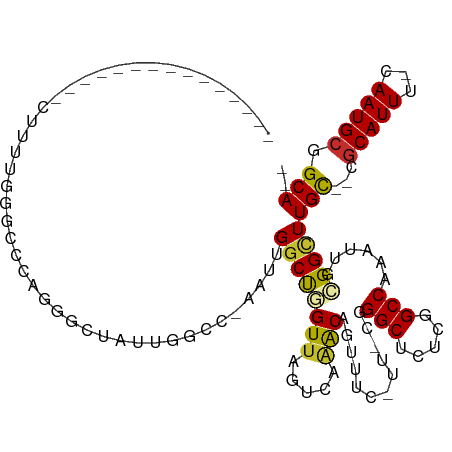
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:56:58 2011