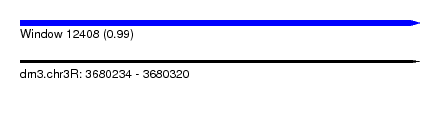
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 3,680,234 – 3,680,320 |
| Length | 86 |
| Max. P | 0.993506 |

| Location | 3,680,234 – 3,680,320 |
|---|---|
| Length | 86 |
| Sequences | 10 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 57.77 |
| Shannon entropy | 0.89977 |
| G+C content | 0.38968 |
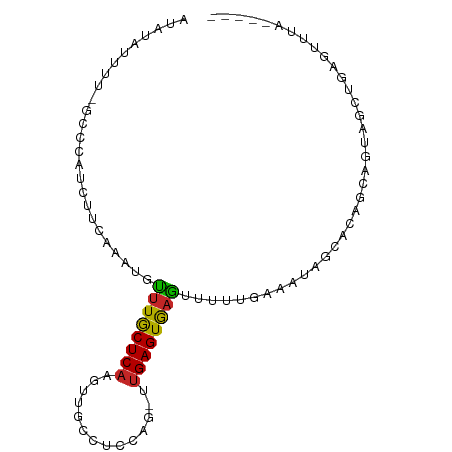
| Mean single sequence MFE | -22.65 |
| Consensus MFE | -5.39 |
| Energy contribution | -5.55 |
| Covariance contribution | 0.16 |
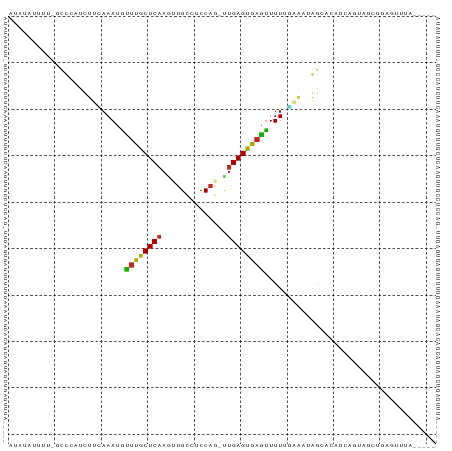
| Combinations/Pair | 1.62 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.24 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.993506 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
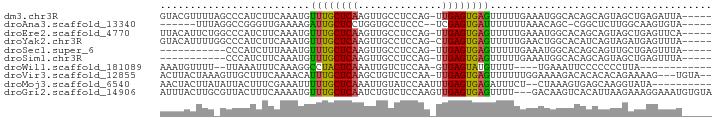
>dm3.chr3R 3680234 86 + 27905053 GUACGUUUUAGCCCAUCUUCAAAUGUUUGCUCAAGUUGCCUCCAG-UUGAGUGAGUUUUUGAAAUGGCACAGCAGUAGCUGAGAUUA----- ....((((((((((((.((((((..((..(((((.(((....)))-)))))..))..)))))))))).((....)).))))))))..----- ( -23.80, z-score = -2.03, R) >droAna3.scaffold_13340 10868004 78 + 23697760 ------UUUAGGCCGGGUUGAAAAGAUUGCUCCUGGUGCCUCCC--UCGAGUGAUUUUUUUAAACAGC-CGGCUCUUGGCAAGUGUA----- ------....((((((.((((((((((..(((..((......))--..)))..))))))))))....)-))))).............----- ( -27.10, z-score = -2.04, R) >droEre2.scaffold_4770 3919148 86 + 17746568 UUACAUUCUGGCCCAUCUUCAAAUGUUUGCUCAAGUUGCCUCCAG-UUGAGUGAGUUUUUGAAAUGGCACAGCAGUAGCUGAGUUCA----- ..........(((.((.((((((..((..(((((.(((....)))-)))))..))..))))))))))).((((....))))......----- ( -23.50, z-score = -1.20, R) >droYak2.chr3R 22221495 86 - 28832112 GUACAUUUUGGCCCAUCUUCAAAUGUUUGCUCAAGUUGCCUCCAG-CUGAGUGAGUUUUUGAACUGGCACAUCAGUAGAUGAGUUUA----- .........(((.((((((((((..((..((((.((((....)))-)))))..))..)))))(((((....)))))))))).)))..----- ( -23.70, z-score = -1.60, R) >droSec1.super_6 3749944 75 + 4358794 -----------CCCAUCUUUAAAUGUUUGCUCAAGUUGCCUCCAG-UUGAGUGAGUUUUUGAAAUGGCACAGCAGUUGCUGAGUUUA----- -----------.((((.((((((..((..(((((.(((....)))-)))))..))..))))))))))..((((....))))......----- ( -21.00, z-score = -2.16, R) >droSim1.chr3R 3715799 75 + 27517382 -----------CCCAUCUUCAAAUGUUUGCUCAAGUUGCCUCCAG-UUGAGUGAGUUUUUGAAAUGGCACAGCAGUAGCUGAGUUUA----- -----------.((((.((((((..((..(((((.(((....)))-)))))..))..))))))))))..((((....))))......----- ( -22.90, z-score = -2.68, R) >droWil1.scaffold_181089 7564031 73 + 12369635 AAAUGUUUU--UUAAAUUUCAAAGGCCUACUCAAAUUGUCUCCAA-GUGAGUAUGUUUU----UGAAAUUCCCCCCCUUA------------ .........--...((((((((((((.((((((..(((....)))-.)))))).)))))----)))))))..........------------ ( -17.80, z-score = -5.41, R) >droVir3.scaffold_12855 6611747 86 + 10161210 ACUUACUAAAGUUGCUUUCAAAACAUUUGCUCAAGCUGUCUCCAA-UUGAGUGAGUUUUUUGGAAAAGACACACACAGAAAAG---UGUA-- ..........(((..((((((((.(((..(((((..((....)).-)))))..))).))))))))..)))..((((......)---))).-- ( -22.50, z-score = -1.93, R) >droMoj3.scaffold_6540 6985364 80 + 34148556 AACUACUUAUAUUACUUUCGAAAUUUUUGCUCAAAUUGUAUCCAAUUUGAGUGAGAUUUCU--CUAAAGUGAGCAAGGUAUA---------- ...(((((...(((((((.((((.(((..(((((((((....)))))))))..))).)).)--).)))))))...)))))..---------- ( -24.30, z-score = -5.12, R) >droGri2.scaffold_14906 2789119 89 + 14172833 AUUUACUUGCGUUACUUUCAAAAUGUUUGCUCAAUCUGUCUCCAAGUUGAGUGAGUUUU---GACAAGUCACAUUAAGAAAGGAAAUGUGUA ........(((((.(((((..((((((..(((((..((....))..)))))..).....---((....)))))))..)))))..)))))... ( -19.90, z-score = -1.30, R) >consensus AUAUAUUUU_GCCCAUCUUCAAAUGUUUGCUCAAGUUGCCUCCAG_UUGAGUGAGUUUUUGAAAUAGCACAGCAGUAGCUGAGUUUA_____ .................(((.((..((((((((..(((....)))..))))))))..)).)))............................. ( -5.39 = -5.55 + 0.16)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:56:57 2011