
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 3,673,557 – 3,673,666 |
| Length | 109 |
| Max. P | 0.876189 |

| Location | 3,673,557 – 3,673,665 |
|---|---|
| Length | 108 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 61.03 |
| Shannon entropy | 0.80003 |
| G+C content | 0.64674 |
| Mean single sequence MFE | -43.79 |
| Consensus MFE | -12.18 |
| Energy contribution | -12.12 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.88 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.28 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.781439 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
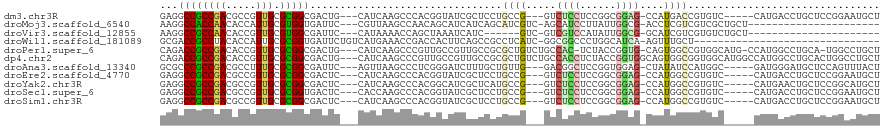
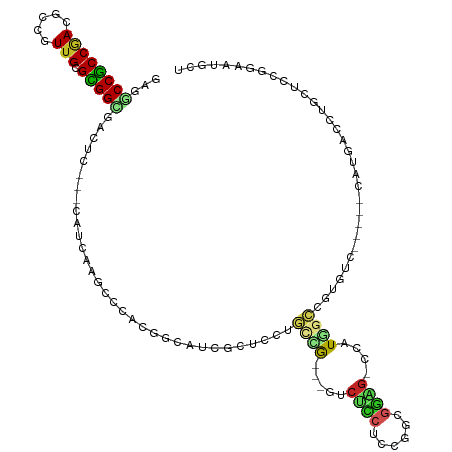
>dm3.chr3R 3673557 108 + 27905053 GAGGCCGCCGACGCCGUUGCGCGGCGACUG---CAUCAAGCCCACGGUAUCGCUCCUGCCG---GUCUCCUCCGGCGGAG-CCAUGACCGUGUC-----CAUGACCUGCUCCGGAAUGCU ..(((..(((.(((((.....)))))...(---(((((.(..((((((...(((((.((((---(......)))))))))-)....))))))..-----).)))..)))..)))...))) ( -50.80, z-score = -2.73, R) >droMoj3.scaffold_6540 6979932 93 + 34148556 AAGGCCACCAACACCAUUGCGUGGUGAUUC---CGUUAAGCCAACAGCAUCAUCAGCAUCGUC-AGCAUCCUUAUUGGCG-ACCUCGUCGUCGCUGCU---------------------- ..(((.((...((((((...))))))....---.))...)))..((((.......((......-.)).........((((-((...))))))))))..---------------------- ( -22.80, z-score = -0.38, R) >droVir3.scaffold_12855 6606165 87 + 10161210 AAGGCCGCCAACACCGUUGCGUGGCGAUUC---CAUAAAACCAGCUAAAUCAUC------GUC-GUCGUCCAUAUUGGCG-GCAUCGUCGUGUCUGCU---------------------- ...(((((((((...)))).))))).....---.........(((.....(..(------(..-((((((......))))-))..))..).....)))---------------------- ( -23.70, z-score = -0.49, R) >droWil1.scaffold_181089 7558809 87 + 12369635 GCGACCGCCUACACCAUUGCGCGGUGAUUCUGUCAUGAAACCGACCACUUCAGCCGCCUCAUC-GGCGGCCCUGGCAUCA-AGUUUGCU------------------------------- (((((((((.........).))))).....(((((((((.........))))((((((.....-))))))..)))))...-....))).------------------------------- ( -25.60, z-score = -0.59, R) >droPer1.super_6 2374197 113 + 6141320 CAGACCGCCGACACCGUUGCGCGGCGACUG---CAUCAAGCCCGUUGCCGUUGCCGCGCUGUCUGCCAC-UCUACCGGUG-CAGUGGCCGUGGCAUG-CCAUGGCCUGCA-UGGCCUGCU (((.((((.((((.....((((((((((.(---((.(......).))).)))))))))))))).))...-.......(((-((..((((((((....-))))))))))))-))).))).. ( -55.60, z-score = -2.17, R) >dp4.chr2 27080795 117 + 30794189 CAGACCGCCGACACCGUUGCGCGGCGACUG---CAUCAAGCCCGUUGCCGUUGCCGCGCUGUCUGCCACCUCUACCGGUGGCAGUGGCGGUGGCAUGGCCAUGGCCUGCACUGGCCUGCU (((...((((.(((((((((((((((((.(---((.(......).))).))))))))))...((((((((......)))))))).)))))))(((.(((....))))))..))))))).. ( -65.20, z-score = -3.38, R) >droAna3.scaffold_13340 10863006 108 + 23697760 GCGCCCGCCGACGCCUUUGCGCGGCGAUUC---AGUUAAGCCCUCGGGAUCUUUGCUGUUG---GACGGCUCCGGUGGAG-CUAUAUCCAUGGC-----GAUGGGAUGCUCCAGUUUACU .(((((((((.(((....))))))))...(---(((.(((((....))..))).)))).((---((.((((((...))))-))...)))).)))-----).((((....))))....... ( -40.20, z-score = -0.23, R) >droEre2.scaffold_4770 3913284 108 + 17746568 GAGGCCGCCGACGCCGUUGCGCGGCGACUC---CAUCAAGCCCACGGUAUCGCUCCUGCCG---GUCUCCUCCGGCGGAG-CCAUGGCCGUGUC-----CAUGACCUGCUCCGGAAUGCU (((..(((((.(((....)))))))).)))---..(((.(..((((((...(((((.((((---(......)))))))))-)....))))))..-----).)))...((........)). ( -50.00, z-score = -2.08, R) >droYak2.chr3R 22214859 108 - 28832112 GAGGCCGCCGACGCCGUUGCGCGGCGACUC---CAUCAAGCCCACGGCAUCGCUCAUGCCG---GUCUCCUCCGGCGGAG-CCAUGGCCGUGUC-----CAUGAACUGCUCCGGCAUGCU (((..(((((.(((....)))))))).)))---..(((.(..((((((...((((.(((((---(......)))))))))-)....))))))..-----).)))..(((....))).... ( -50.60, z-score = -1.82, R) >droSec1.super_6 3744330 108 + 4358794 GAGGCCGCCGACGCCGUUGCGCGGUGACUC---CACCAAGCCCACGGUAUCGCUCCUGCCG---GUCUCCUCCGGCGGAG-CCAUGGCCGUGUC-----CAUGACCUGCUCCGGAAUGCU ((.((.(((((..((((.((..((((....---))))..))..))))..))(((((.((((---(......)))))))))-)...))).)).))-----(((..((......)).))).. ( -47.20, z-score = -1.52, R) >droSim1.chr3R 3710156 108 + 27517382 GAGGCCGCCGACGCCGUUGCGCGGCGACUC---CAUCAAGCCCACGGUAUCGCUCCUGCCG---GUCUCCUCCGGCGGAG-CCAUGGCCGUGUC-----CAUGACCUGCUCCGGAAUGCU (((..(((((.(((....)))))))).)))---..(((.(..((((((...(((((.((((---(......)))))))))-)....))))))..-----).)))...((........)). ( -50.00, z-score = -2.08, R) >consensus GAGGCCGCCGACGCCGUUGCGCGGCGACUC___CAUCAAGCCCACGGCAUCGCUCCUGCCG___GUCUCCUCCGGCGGAG_CCAUGGCCGUGUC_____CAUGACCUGCUCCGGAAUGCU ...((((((((.....))).)))))................................((((.....((((......))))....))))................................ (-12.18 = -12.12 + -0.06)
| Location | 3,673,557 – 3,673,665 |
|---|---|
| Length | 108 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 61.03 |
| Shannon entropy | 0.80003 |
| G+C content | 0.64674 |
| Mean single sequence MFE | -47.66 |
| Consensus MFE | -12.94 |
| Energy contribution | -12.66 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.856134 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
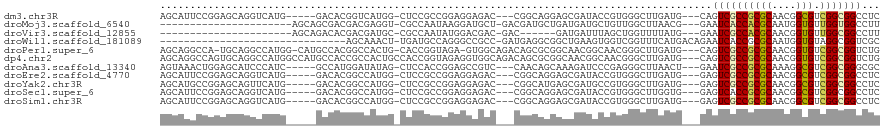
>dm3.chr3R 3673557 108 - 27905053 AGCAUUCCGGAGCAGGUCAUG-----GACACGGUCAUGG-CUCCGCCGGAGGAGAC---CGGCAGGAGCGAUACCGUGGGCUUGAUG---CAGUCGCCGCGCAACGGCGUCGGCGGCCUC .(((((...((((........-----..((((((.((.(-(((((((((......)---)))).))))).)))))))).))))))))---).((((((((((....))).)))))))... ( -56.40, z-score = -2.96, R) >droMoj3.scaffold_6540 6979932 93 - 34148556 ----------------------AGCAGCGACGACGAGGU-CGCCAAUAAGGAUGCU-GACGAUGCUGAUGAUGCUGUUGGCUUAACG---GAAUCACCACGCAAUGGUGUUGGUGGCCUU ----------------------............(((((-((((((((..(.((((-(..(((.(((.(((.((.....))))).))---).)))..)).))).)..))))))))))))) ( -30.30, z-score = -0.95, R) >droVir3.scaffold_12855 6606165 87 - 10161210 ----------------------AGCAGACACGACGAUGC-CGCCAAUAUGGACGAC-GAC------GAUGAUUUAGCUGGUUUUAUG---GAAUCGCCACGCAACGGUGUUGGCGGCCUU ----------------------...............((-(((((((((.(.((..-..)------)........((((((......---.....)))).))..).)))))))))))... ( -25.90, z-score = -0.79, R) >droWil1.scaffold_181089 7558809 87 - 12369635 -------------------------------AGCAAACU-UGAUGCCAGGGCCGCC-GAUGAGGCGGCUGAAGUGGUCGGUUUCAUGACAGAAUCACCGCGCAAUGGUGUAGGCGGUCGC -------------------------------.((..(((-...(((...(((((((-.....)))))))...(((((.(((((.......)))))))))))))..)))....))...... ( -31.20, z-score = -0.47, R) >droPer1.super_6 2374197 113 - 6141320 AGCAGGCCA-UGCAGGCCAUGG-CAUGCCACGGCCACUG-CACCGGUAGA-GUGGCAGACAGCGCGGCAACGGCAACGGGCUUGAUG---CAGUCGCCGCGCAACGGUGUCGGCGGUCUG ..((((((.-....((((.(((-....))).)))).(((-(((((((...-...)).....(((((((..((....))((((.....---.)))))))))))..))))).))).)))))) ( -56.30, z-score = -1.53, R) >dp4.chr2 27080795 117 - 30794189 AGCAGGCCAGUGCAGGCCAUGGCCAUGCCACCGCCACUGCCACCGGUAGAGGUGGCAGACAGCGCGGCAACGGCAACGGGCUUGAUG---CAGUCGCCGCGCAACGGUGUCGGCGGUCUG ..((((((......(((....)))..(((..((((.((((((((......))))))))...(((((((..((....))((((.....---.)))))))))))...))))..))))))))) ( -68.00, z-score = -3.81, R) >droAna3.scaffold_13340 10863006 108 - 23697760 AGUAAACUGGAGCAUCCCAUC-----GCCAUGGAUAUAG-CUCCACCGGAGCCGUC---CAACAGCAAAGAUCCCGAGGGCUUAACU---GAAUCGCCGCGCAAAGGCGUCGGCGGGCGC .......(((......))).(-----(((.(((((...(-((((...))))).)))---)).(((..(((.(((...))))))..))---)...((((((((....))).))))))))). ( -38.70, z-score = -0.93, R) >droEre2.scaffold_4770 3913284 108 - 17746568 AGCAUUCCGGAGCAGGUCAUG-----GACACGGCCAUGG-CUCCGCCGGAGGAGAC---CGGCAGGAGCGAUACCGUGGGCUUGAUG---GAGUCGCCGCGCAACGGCGUCGGCGGCCUC .....((((...((((((...-----..(((((..((.(-(((((((((......)---)))).))))).)).))))))))))).))---))((((((((((....))).)))))))... ( -56.40, z-score = -2.71, R) >droYak2.chr3R 22214859 108 + 28832112 AGCAUGCCGGAGCAGUUCAUG-----GACACGGCCAUGG-CUCCGCCGGAGGAGAC---CGGCAUGAGCGAUGCCGUGGGCUUGAUG---GAGUCGCCGCGCAACGGCGUCGGCGGCCUC ..((.((((((....)))...-----..((((((.((.(-(((.(((((......)---))))..)))).))))))))))).))..(---..((((((((((....))).)))))))..) ( -53.10, z-score = -1.30, R) >droSec1.super_6 3744330 108 - 4358794 AGCAUUCCGGAGCAGGUCAUG-----GACACGGCCAUGG-CUCCGCCGGAGGAGAC---CGGCAGGAGCGAUACCGUGGGCUUGGUG---GAGUCACCGCGCAACGGCGUCGGCGGCCUC .((.(((((((((.((((...-----.....))))...)-))))(((((......)---)))).))))(((..((((..((.(((((---....))))).)).))))..)))))...... ( -51.60, z-score = -1.47, R) >droSim1.chr3R 3710156 108 - 27517382 AGCAUUCCGGAGCAGGUCAUG-----GACACGGCCAUGG-CUCCGCCGGAGGAGAC---CGGCAGGAGCGAUACCGUGGGCUUGAUG---GAGUCGCCGCGCAACGGCGUCGGCGGCCUC .....((((...((((((...-----..(((((..((.(-(((((((((......)---)))).))))).)).))))))))))).))---))((((((((((....))).)))))))... ( -56.40, z-score = -2.71, R) >consensus AGCAUUCCGGAGCAGGUCAUG_____GACACGGCCAUGG_CUCCGCCGGAGGAGAC___CGGCAGGAGCGAUGCCGUGGGCUUGAUG___GAGUCGCCGCGCAACGGCGUCGGCGGCCUC ............................................................................................((((((((((....))).)))))))... (-12.94 = -12.66 + -0.28)
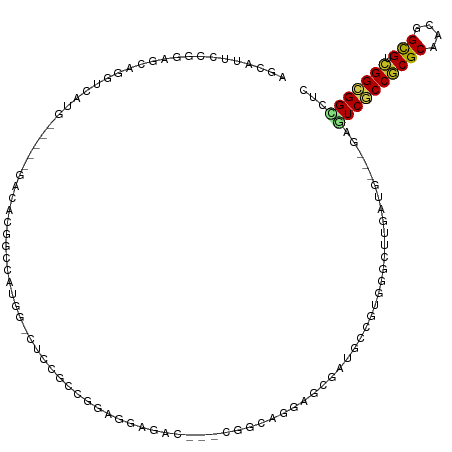
| Location | 3,673,558 – 3,673,666 |
|---|---|
| Length | 108 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 60.96 |
| Shannon entropy | 0.79772 |
| G+C content | 0.64718 |
| Mean single sequence MFE | -43.59 |
| Consensus MFE | -11.64 |
| Energy contribution | -11.57 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.94 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.807458 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
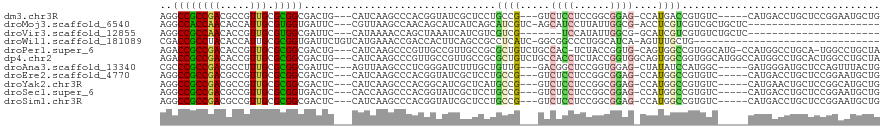
>dm3.chr3R 3673558 108 + 27905053 AGGCCGCCGACGCCGUUGCGCGGCGACUG---CAUCAAGCCCACGGUAUCGCUCCUGCCG---GUCUCCUCCGGCGGAG-CCAUGACCGUGUC-----CAUGACCUGCUCCGGAAUGCUG .(((..(((.(((((.....)))))...(---(((((.(..((((((...(((((.((((---(......)))))))))-)....))))))..-----).)))..)))..)))...))). ( -51.50, z-score = -3.11, R) >droMoj3.scaffold_6540 6979933 93 + 34148556 AGGCCACCAACACCAUUGCGUGGUGAUUC---CGUUAAGCCAACAGCAUCAUCAGCAUCGUC-AGCAUCCUUAUUGGCG-ACCUCGUCGUCGCUGCUC---------------------- .(((.((...((((((...))))))....---.))...)))..((((.......((......-.)).........((((-((...))))))))))...---------------------- ( -22.80, z-score = -0.43, R) >droVir3.scaffold_12855 6606166 87 + 10161210 AGGCCGCCAACACCGUUGCGUGGCGAUUC---CAUAAAACCAGCUAAAUCAUCGUCGUCG-------UCCAUAUUGGCG-GCAUCGUCGUGUCUGCUC---------------------- ..(((((((((...)))).))))).....---.........(((.....(..((..((((-------((......))))-))..))..).....))).---------------------- ( -23.80, z-score = -0.58, R) >droWil1.scaffold_181089 7558810 87 + 12369635 CGACCGCCUACACCAUUGCGCGGUGAUUCUGUCAUGAAACCGACCACUUCAGCCGCCUCAUC-GGCGGCCCUGGCAUCA-AGUUUGCUG------------------------------- ..((((((.........).)))))((....(((........)))....)).((((((.....-))))))...((((...-....)))).------------------------------- ( -25.10, z-score = -0.71, R) >droPer1.super_6 2374198 113 + 6141320 AGACCGCCGACACCGUUGCGCGGCGACUG---CAUCAAGCCCGUUGCCGUUGCCGCGCUGUCUGCCAC-UCUACCGGUG-CAGUGGCCGUGGCAUG-CCAUGGCCUGCA-UGGCCUGCUA ((...(((((((.....((((((((((.(---((.(......).))).))))))))))))))......-.......(((-((..((((((((....-))))))))))))-))))...)). ( -53.60, z-score = -1.76, R) >dp4.chr2 27080796 117 + 30794189 AGACCGCCGACACCGUUGCGCGGCGACUG---CAUCAAGCCCGUUGCCGUUGCCGCGCUGUCUGCCACCUCUACCGGUGGCAGUGGCGGUGGCAUGGCCAUGGCCUGCACUGGCCUGCUA ..(((((((........((((((((((.(---((.(......).))).))))))))))...((((((((......)))))))))))))))((((.(((((((.....)).))))))))). ( -65.90, z-score = -3.71, R) >droAna3.scaffold_13340 10863007 108 + 23697760 CGCCCGCCGACGCCUUUGCGCGGCGAUUC---AGUUAAGCCCUCGGGAUCUUUGCUGUUG---GACGGCUCCGGUGGAG-CUAUAUCCAUGGC-----GAUGGGAUGCUCCAGUUUACUG (((((((((.(((....))))))))...(---(((.(((((....))..))).)))).((---((.((((((...))))-))...)))).)))-----).((((....))))........ ( -40.00, z-score = -0.51, R) >droEre2.scaffold_4770 3913285 108 + 17746568 AGGCCGCCGACGCCGUUGCGCGGCGACUC---CAUCAAGCCCACGGUAUCGCUCCUGCCG---GUCUCCUCCGGCGGAG-CCAUGGCCGUGUC-----CAUGACCUGCUCCGGAAUGCUG .(((..(((.(((((.....)))))....---..(((.(..((((((...(((((.((((---(......)))))))))-)....))))))..-----).))).......)))...))). ( -49.20, z-score = -2.04, R) >droYak2.chr3R 22214860 108 - 28832112 AGGCCGCCGACGCCGUUGCGCGGCGACUC---CAUCAAGCCCACGGCAUCGCUCAUGCCG---GUCUCCUCCGGCGGAG-CCAUGGCCGUGUC-----CAUGAACUGCUCCGGCAUGCUG .(((.((((.(((((.....)))))....---..(((.(..((((((...((((.(((((---(......)))))))))-)....))))))..-----).))).......))))..))). ( -51.30, z-score = -2.18, R) >droSec1.super_6 3744331 108 + 4358794 AGGCCGCCGACGCCGUUGCGCGGUGACUC---CACCAAGCCCACGGUAUCGCUCCUGCCG---GUCUCCUCCGGCGGAG-CCAUGGCCGUGUC-----CAUGACCUGCUCCGGAAUGCUG .(((((..((..((((.((..((((....---))))..))..))))..))(((((.((((---(......)))))))))-)..)))))(((((-----(............)).)))).. ( -47.10, z-score = -1.66, R) >droSim1.chr3R 3710157 108 + 27517382 AGGCCGCCGACGCCGUUGCGCGGCGACUC---CAUCAAGCCCACGGUAUCGCUCCUGCCG---GUCUCCUCCGGCGGAG-CCAUGGCCGUGUC-----CAUGACCUGCUCCGGAAUGCUG .(((..(((.(((((.....)))))....---..(((.(..((((((...(((((.((((---(......)))))))))-)....))))))..-----).))).......)))...))). ( -49.20, z-score = -2.04, R) >consensus AGGCCGCCGACGCCGUUGCGCGGCGACUC___CAUCAAGCCCACGGCAUCGCUCCUGCCG___GUCUCCUCCGGCGGAG_CCAUGGCCGUGUC_____CAUGACCUGCUCCGGAAUGCUG ..((((((((.....))).)))))................................((((.....((((......))))....))))................................. (-11.64 = -11.57 + -0.07)

| Location | 3,673,558 – 3,673,666 |
|---|---|
| Length | 108 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 60.96 |
| Shannon entropy | 0.79772 |
| G+C content | 0.64718 |
| Mean single sequence MFE | -47.23 |
| Consensus MFE | -12.94 |
| Energy contribution | -12.66 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.876189 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 3673558 108 - 27905053 CAGCAUUCCGGAGCAGGUCAUG-----GACACGGUCAUGG-CUCCGCCGGAGGAGAC---CGGCAGGAGCGAUACCGUGGGCUUGAUG---CAGUCGCCGCGCAACGGCGUCGGCGGCCU ..(((((...((((........-----..((((((.((.(-(((((((((......)---)))).))))).)))))))).))))))))---).((((((((((....))).))))))).. ( -56.40, z-score = -3.07, R) >droMoj3.scaffold_6540 6979933 93 - 34148556 ----------------------GAGCAGCGACGACGAGGU-CGCCAAUAAGGAUGCU-GACGAUGCUGAUGAUGCUGUUGGCUUAACG---GAAUCACCACGCAAUGGUGUUGGUGGCCU ----------------------..............((((-((((((((..(.((((-(..(((.(((.(((.((.....))))).))---).)))..)).))).)..)))))))))))) ( -29.10, z-score = -0.60, R) >droVir3.scaffold_12855 6606166 87 - 10161210 ----------------------GAGCAGACACGACGAUGC-CGCCAAUAUGGA-------CGACGACGAUGAUUUAGCUGGUUUUAUG---GAAUCGCCACGCAACGGUGUUGGCGGCCU ----------------------................((-(((((((((.(.-------((....))........((((((......---.....)))).))..).))))))))))).. ( -25.90, z-score = -0.79, R) >droWil1.scaffold_181089 7558810 87 - 12369635 -------------------------------CAGCAAACU-UGAUGCCAGGGCCGCC-GAUGAGGCGGCUGAAGUGGUCGGUUUCAUGACAGAAUCACCGCGCAAUGGUGUAGGCGGUCG -------------------------------..((..(((-...(((...(((((((-.....)))))))...(((((.(((((.......)))))))))))))..)))....))..... ( -31.20, z-score = -0.68, R) >droPer1.super_6 2374198 113 - 6141320 UAGCAGGCCA-UGCAGGCCAUGG-CAUGCCACGGCCACUG-CACCGGUAGA-GUGGCAGACAGCGCGGCAACGGCAACGGGCUUGAUG---CAGUCGCCGCGCAACGGUGUCGGCGGUCU ..(((.((((-((.....)))))-).)))...((((.(((-(((((((...-...)).....(((((((..((....))((((.....---.)))))))))))..))))).))).)))). ( -55.00, z-score = -1.52, R) >dp4.chr2 27080796 117 - 30794189 UAGCAGGCCAGUGCAGGCCAUGGCCAUGCCACCGCCACUGCCACCGGUAGAGGUGGCAGACAGCGCGGCAACGGCAACGGGCUUGAUG---CAGUCGCCGCGCAACGGUGUCGGCGGUCU ....(((((......(((....)))..(((..((((.((((((((......))))))))...(((((((..((....))((((.....---.)))))))))))...))))..)))))))) ( -65.50, z-score = -3.45, R) >droAna3.scaffold_13340 10863007 108 - 23697760 CAGUAAACUGGAGCAUCCCAUC-----GCCAUGGAUAUAG-CUCCACCGGAGCCGUC---CAACAGCAAAGAUCCCGAGGGCUUAACU---GAAUCGCCGCGCAAAGGCGUCGGCGGGCG (((....)))...........(-----(((.(((((...(-((((...))))).)))---)).(((..(((.(((...))))))..))---)...((((((((....))).))))))))) ( -38.40, z-score = -1.25, R) >droEre2.scaffold_4770 3913285 108 - 17746568 CAGCAUUCCGGAGCAGGUCAUG-----GACACGGCCAUGG-CUCCGCCGGAGGAGAC---CGGCAGGAGCGAUACCGUGGGCUUGAUG---GAGUCGCCGCGCAACGGCGUCGGCGGCCU ......((((...((((((...-----..(((((..((.(-(((((((((......)---)))).))))).)).))))))))))).))---))((((((((((....))).))))))).. ( -56.40, z-score = -2.85, R) >droYak2.chr3R 22214860 108 + 28832112 CAGCAUGCCGGAGCAGUUCAUG-----GACACGGCCAUGG-CUCCGCCGGAGGAGAC---CGGCAUGAGCGAUGCCGUGGGCUUGAUG---GAGUCGCCGCGCAACGGCGUCGGCGGCCU ((.((.((((((....)))...-----..((((((.((.(-(((.(((((......)---))))..)))).))))))))))).)).))---..((((((((((....))).))))))).. ( -53.60, z-score = -1.51, R) >droSec1.super_6 3744331 108 - 4358794 CAGCAUUCCGGAGCAGGUCAUG-----GACACGGCCAUGG-CUCCGCCGGAGGAGAC---CGGCAGGAGCGAUACCGUGGGCUUGGUG---GAGUCACCGCGCAACGGCGUCGGCGGCCU ..((.(((((((((.((((...-----.....))))...)-))))(((((......)---)))).))))(((..((((..((.(((((---....))))).)).))))..)))))..... ( -51.60, z-score = -1.59, R) >droSim1.chr3R 3710157 108 - 27517382 CAGCAUUCCGGAGCAGGUCAUG-----GACACGGCCAUGG-CUCCGCCGGAGGAGAC---CGGCAGGAGCGAUACCGUGGGCUUGAUG---GAGUCGCCGCGCAACGGCGUCGGCGGCCU ......((((...((((((...-----..(((((..((.(-(((((((((......)---)))).))))).)).))))))))))).))---))((((((((((....))).))))))).. ( -56.40, z-score = -2.85, R) >consensus CAGCAUUCCGGAGCAGGUCAUG_____GACACGGCCAUGG_CUCCGCCGGAGGAGAC___CGGCAGGAGCGAUGCCGUGGGCUUGAUG___GAGUCGCCGCGCAACGGCGUCGGCGGCCU .............................................................................................((((((((((....))).))))))).. (-12.94 = -12.66 + -0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:56:56 2011