
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 3,572,072 – 3,572,169 |
| Length | 97 |
| Max. P | 0.582010 |

| Location | 3,572,072 – 3,572,169 |
|---|---|
| Length | 97 |
| Sequences | 11 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 72.35 |
| Shannon entropy | 0.58125 |
| G+C content | 0.38792 |
| Mean single sequence MFE | -21.47 |
| Consensus MFE | -8.02 |
| Energy contribution | -8.47 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.582010 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 3572072 97 + 27905053 -----UCUGCC-AUUUUAUAGCCACUUUAUCUGUUUAAUUAAAAGCCUGGCUUUUAAUUGAAAUCGCUCU----GAUAUCUAAGCUGUCACAAUUGCCAUCUGGCCC---- -----......-.....(((((.....((((.((((((((((((((...))))))))))))....))...----)))).....))))).......(((....)))..---- ( -19.30, z-score = -1.66, R) >droSim1.chr3R 3607789 97 + 27517382 -----UCUGCC-AUUUUAUAGCCACUUUAUCUGUUUAAUUAAAAGCCUGGCUUUUAAUUGAAAUCGCUCU----GAUAUCUGAGCUGUCACAAUUGCCAUCUGGCCC---- -----......-........((((.........(((((((((((((...)))))))))))))...((((.----.......))))................))))..---- ( -21.50, z-score = -2.10, R) >droSec1.super_6 3636745 97 + 4358794 -----UCUGCC-AUUUUAUAGCCACUUUAUCUGUUUAAUUAAAAGCCUGGCUUUUAAUUGAAAUCGCUCU----GAUAUCUGAGCUGUCACAAUUGCCAUCUGGCCC---- -----......-........((((.........(((((((((((((...)))))))))))))...((((.----.......))))................))))..---- ( -21.50, z-score = -2.10, R) >droYak2.chr3R_random 381264 97 + 3277457 -----UCUCUC-AUUUUAUAGCCAUUUUAUCUGUUUAAUUAAAAGGCUGGCUUUUAAUUGAAAUCGCUCU----GAUAUCUGAGCUGUCACAAUUGCCAUCUGGCCC---- -----......-........((((.........((((((((((((.....))))))))))))...((((.----.......))))................))))..---- ( -19.70, z-score = -1.33, R) >droEre2.scaffold_4770 3810942 97 + 17746568 -----UCUCCC-ACUUUAUAGCCACUUUAUCUGUUUAAUUAAAAGGCUGGCUUUUAAUUGAAAUCGCUCU----GAUAUCUGAGCUGUCACAAUUGCCAUCUGGCCC---- -----......-........((((.........((((((((((((.....))))))))))))...((((.----.......))))................))))..---- ( -19.60, z-score = -1.40, R) >droAna3.scaffold_13340 3310497 100 + 23697760 ---CCUCUAUCCAUUUCAUAGCCACCUUAUCUGCUUAAUUAAAUGGUUGGCCUUUAAUUGAAAUCGCUCC----GAUAUCUGACCUGUCACAAUUUCCAGCAGGCCU---- ---.......((((((...(((..........))).....))))))..(((((..(((((..((((...)----)))...(((....))))))))......))))).---- ( -16.30, z-score = -0.47, R) >droPer1.super_6 2272057 108 + 6141320 CUUGCUCUAUCAUUUUAUGGGGCACUUUAUCUGUUUAAUUAAAAGGUUGGCUCUUAAUUGAAAUCGCUUCU---GAUAUUUGAGCUGUCACAAUUGCCAGCUGACCUGGCC ..((((((((......))))))))...................((((..(((..((((((..((.(((.(.---.......)))).))..))))))..)))..)))).... ( -27.80, z-score = -1.88, R) >dp4.chr2 26966438 108 + 30794189 CUUGCUCUAUCAUUUUAUGGGGCACUUUAUCUGUUUAAUUAAAAGGUUGGCUCUUAAUUGAAAUCGCUUCU---GAUAUUUGAGCUGUCACAAUUGCCAGCUGACCUGGCC ..((((((((......))))))))...................((((..(((..((((((..((.(((.(.---.......)))).))..))))))..)))..)))).... ( -27.80, z-score = -1.88, R) >droWil1.scaffold_181089 4921624 92 - 12369635 -AUGACUUUUAGAUCUUAUGGCCAUUUAAUCUGUUUAAUUAAAUG-CUGGCAUUUAAUUGGAUACAUUCUACCACAUACUUGAGAUGUCGAAAU----------------- -.((((((((((....((((.......(((.((((((((((((((-....)))))))))))))).)))......)))).)))))).))))....----------------- ( -18.12, z-score = -1.84, R) >droMoj3.scaffold_6540 6839936 95 + 34148556 --------GCCGUUUUUAUGGCCACUUAAUCUGUUUAAUUAAAAGUCUGGCAUUUGAUCGCAAAUGCUUU----AAUAUUCGAGCCGGGCCGGUUGUUAAGUGUCUC---- --------(((((....)))))((((((((..................((((((((....))))))))..----........(((((...)))))))))))))....---- ( -24.30, z-score = -1.73, R) >droGri2.scaffold_14906 6540939 93 - 14172833 --------GACGCUUUUAUGACCACUUAAUCGCUUUAAUUAAAAGUCGGCCAUCUAAUCAAGAAUGUUCU----GAUAUUUGAGCUGCCACAAUGAGCGUCUAGC------ --------(((((((...((...........(((((.....))))).(((..(((.....)))..((((.----.......)))).))).))..)))))))....------ ( -20.30, z-score = -1.33, R) >consensus _____UCUGCC_AUUUUAUAGCCACUUUAUCUGUUUAAUUAAAAGGCUGGCUUUUAAUUGAAAUCGCUCU____GAUAUCUGAGCUGUCACAAUUGCCAUCUGGCCC____ .................................((((((((((((.....))))))))))))...((((............)))).......................... ( -8.02 = -8.47 + 0.45)

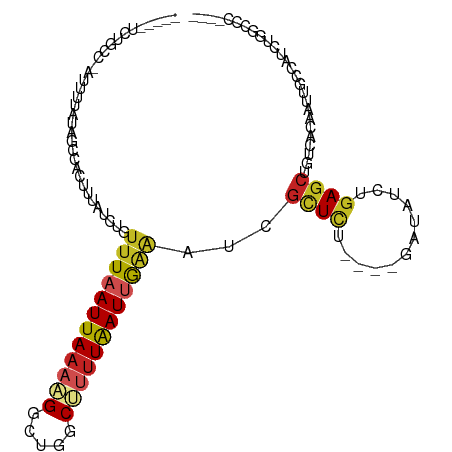
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:56:45 2011