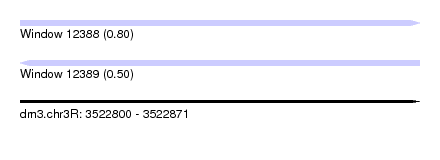
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 3,522,800 – 3,522,871 |
| Length | 71 |
| Max. P | 0.796715 |

| Location | 3,522,800 – 3,522,871 |
|---|---|
| Length | 71 |
| Sequences | 3 |
| Columns | 87 |
| Reading direction | forward |
| Mean pairwise identity | 66.28 |
| Shannon entropy | 0.46920 |
| G+C content | 0.55993 |
| Mean single sequence MFE | -29.00 |
| Consensus MFE | -12.86 |
| Energy contribution | -13.33 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.796715 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 3522800 71 + 27905053 CAAUUCAAGAUCUGGCC-----------GGCCAGGUCGCGGCCAU-ACAGAUUGCCCCUGCCCGG----GCAGAAGCUCCCCCAAUC ..........((((.((-----------((.((((..(((..(..-...)..))).)))).))))----.))))............. ( -23.80, z-score = -0.80, R) >dp4.Unknown_group_92 4842 87 - 26366 CACUGCAAAAUCAGGCCUCGUGGUUUGAGUCCAGGUUACUGUAAUUACAAGCCGCGAUUGCUCAGUUGAGCACAAGCACGCCUGCCA ...........(((((.(((((((((((((.(((....)))..))).)))))))))).(((((....))))).......)))))... ( -30.90, z-score = -2.40, R) >droPer1.super_143 21433 87 + 108547 CACUGCAAAAUCAGGCCUCGCGGUUUGAGACCAGGUUACUGUAAUUACAAGCCGCGAUUGCUCAGUUGAGCACAAGCACGCCUGCCA ...........(((((.((((((((((.((.(((....)))...)).)))))))))).(((((....))))).......)))))... ( -32.30, z-score = -2.91, R) >consensus CACUGCAAAAUCAGGCCUCG_GGUUUGAGACCAGGUUACUGUAAUUACAAGCCGCGAUUGCUCAGUUGAGCACAAGCACGCCUGCCA ........((((.((.((((.....)))).)).))))................((...(((((....)))))...)).......... (-12.86 = -13.33 + 0.47)

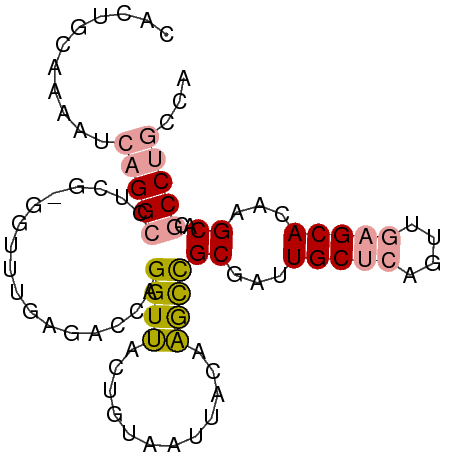

| Location | 3,522,800 – 3,522,871 |
|---|---|
| Length | 71 |
| Sequences | 3 |
| Columns | 87 |
| Reading direction | reverse |
| Mean pairwise identity | 66.28 |
| Shannon entropy | 0.46920 |
| G+C content | 0.55993 |
| Mean single sequence MFE | -29.10 |
| Consensus MFE | -17.83 |
| Energy contribution | -18.90 |
| Covariance contribution | 1.07 |
| Combinations/Pair | 1.29 |
| Mean z-score | -0.99 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr3R 3522800 71 - 27905053 GAUUGGGGGAGCUUCUGC----CCGGGCAGGGGCAAUCUGU-AUGGCCGCGACCUGGCC-----------GGCCAGAUCUUGAAUUG ((((.((((.((((((((----....))))))))..((((.-.((((((.....)))))-----------)..)))))))).)))). ( -32.00, z-score = -1.69, R) >dp4.Unknown_group_92 4842 87 + 26366 UGGCAGGCGUGCUUGUGCUCAACUGAGCAAUCGCGGCUUGUAAUUACAGUAACCUGGACUCAAACCACGAGGCCUGAUUUUGCAGUG ..((((((.(((...(((((....)))))...))))))))).......((((...((.(((.......))).)).....)))).... ( -25.00, z-score = -0.01, R) >droPer1.super_143 21433 87 - 108547 UGGCAGGCGUGCUUGUGCUCAACUGAGCAAUCGCGGCUUGUAAUUACAGUAACCUGGUCUCAAACCGCGAGGCCUGAUUUUGCAGUG ...(((((.......(((((....))))).((((((.(((......(((....)))....))).)))))).)))))........... ( -30.30, z-score = -1.27, R) >consensus UGGCAGGCGUGCUUGUGCUCAACUGAGCAAUCGCGGCUUGUAAUUACAGUAACCUGGACUCAAACC_CGAGGCCUGAUUUUGCAGUG ..((((((.(((...(((((....)))))...))))))))).......((((...((((((.......)))))).....)))).... (-17.83 = -18.90 + 1.07)


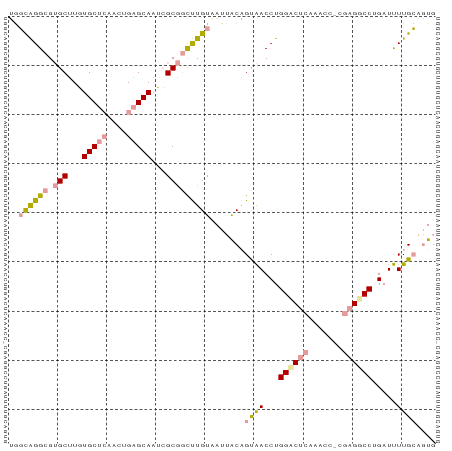
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:56:41 2011