| Sequence ID | dm3.chr3R |
|---|---|
| Location | 3,500,291 – 3,500,383 |
| Length | 92 |
| Max. P | 0.977103 |

| Location | 3,500,291 – 3,500,383 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 63.99 |
| Shannon entropy | 0.70772 |
| G+C content | 0.53075 |
| Mean single sequence MFE | -28.63 |
| Consensus MFE | -12.59 |
| Energy contribution | -12.43 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.977103 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
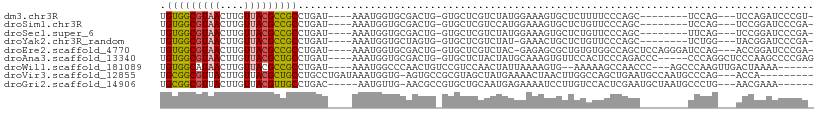
>dm3.chr3R 3500291 92 - 27905053 UGUGGCGUAACUUGUUACGCCGCCUGAU----AAAUGGUGCGACUG-GUGCUCGUCUAUGGAAAGUGCUCUUUUCCCAGC--------UCCAG---UCCAGAUCCCGU- .(((((((((....))))))))).....----..((((((.(((((-(.(((.(.....((((((....))))))).)))--------.))))---))))....))))- ( -32.90, z-score = -3.24, R) >droSim1.chr3R 3561655 92 - 27517382 UGUGGCGUAACUUGUUACGCCGCCUGAU----AAAUGGUGCGACUG-GUGCUCGUCCAUGGAAAGUGCUCUGUUCCCAGC--------UCCAG---UCCGGAUCCCGA- .(((((((((....))))))))).....----....(((.((((((-(.(((.(...(((((......)))))..).)))--------.))))---).)).)))....- ( -31.90, z-score = -2.02, R) >droSec1.super_6 3591555 92 - 4358794 UGUGGCGUAACUUGUUACGCCGCCUGAU----AAAUGGUGCGACUG-GUGCUCGUCUAUGGAAAGUGCUCUGUUCCCAGC--------UUCAG---UCCGGAUCCCGA- .(((((((((....))))))))).....----....(((.((((((-(.(((.(...(((((......)))))..).)))--------.))))---).)).)))....- ( -29.40, z-score = -1.40, R) >droYak2.chr3R_random 1095511 91 - 3277457 UGUGGCGUAACUUGUUACGCCGCCUGAU----AAAUGGUGCUAGUG-GUGCUCGUCUAU-GAAACUGCUCUGUUCCCAGC--------UCUGG---UACGGAUCCCGA- .(((((((((....))))))))).....----...(((.....(..-((..(((....)-)).))..)(((((..(((..--------..)))---.)))))..))).- ( -26.90, z-score = -1.16, R) >droEre2.scaffold_4770 3760172 99 - 17746568 UGUGGCGUAACUUGUUACGCCGCCUGAU----AAAUGGUGCGACUG-GUGCUCGUCUAC-GAGAGCGCUGUGUGGCCAGCUCCAGGGAUCCAG---ACCGGAUCCCGA- .(((((((((....))))))))).....----...(((.((.((((-((((((......-..)))))))).)).)).....)))(((((((..---...)))))))..- ( -45.50, z-score = -3.55, R) >droAna3.scaffold_13340 3266589 99 - 23697760 UGUGGCGUAACUUGUUACGCUGCCUGAU----AAAUGGUGCGACUG-GUGCUCUACUAUGCAAAGUGUUCCACUCCCAGACCC-----CCCAGGCUCCCAAGCCCCGAG .(..((((((....))))))..)(((..----...(((.((.(((.-.(((........))).))))).)))....)))....-----....((((....))))..... ( -27.10, z-score = -0.99, R) >droWil1.scaffold_181089 7402356 94 - 12369635 UGUGGCAUAACUUGUUACGCCGCCUGAU----AAAUGGCCCAACUGUCCGUCCAACUAUUAAAAGUG--AAAAAGCCAACCC---AGCCCAAGUUGACUAAAA------ .(((((.(((....))).))))).....----...((((...(((..................))).--.....))))...(---(((....)))).......------ ( -14.87, z-score = 0.08, R) >droVir3.scaffold_12855 6426364 96 - 10161210 UGCGGCGUUACUUGUUACGCUGCCUGCCUGAUAAAUGGUG-AGUGCCGCGUAGCUAUGAAAACUAACUUGGCCAGCUGAAUGCCAAUGCCCAG---ACCA--------- ((.((((((....(((((((.(((((((........))).-)).)).)))))))...............(((.........))))))))))).---....--------- ( -28.60, z-score = -1.16, R) >droGri2.scaffold_14906 6496570 94 + 14172833 UGCGGCGUUACUUGUUACGUUGCCUGAC-----AAUGUUG-AACGCCGUGCUGCAAUGAGAAAAUCCUUGUCCACUCGAAUGCUAAUGCCCUG---AACGAAA------ .((((((((.......((((((.....)-----)))))..-))))))))((.(((.((((..............))))..)))....))....---.......------ ( -20.54, z-score = -0.90, R) >consensus UGUGGCGUAACUUGUUACGCCGCCUGAU____AAAUGGUGCGACUG_GUGCUCGUCUAUGGAAAGUGCUCUGUUCCCAGC________UCCAG___ACCGGAUCCCGA_ .(((((((((....)))))))))...................................................................................... (-12.59 = -12.43 + -0.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:56:40 2011