| Sequence ID | dm3.chr3R |
|---|---|
| Location | 3,452,658 – 3,452,754 |
| Length | 96 |
| Max. P | 0.978418 |

| Location | 3,452,658 – 3,452,754 |
|---|---|
| Length | 96 |
| Sequences | 8 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 72.30 |
| Shannon entropy | 0.52225 |
| G+C content | 0.51882 |
| Mean single sequence MFE | -25.49 |
| Consensus MFE | -8.94 |
| Energy contribution | -10.24 |
| Covariance contribution | 1.30 |
| Combinations/Pair | 1.37 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.00 |
| SVM RNA-class probability | 0.978418 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
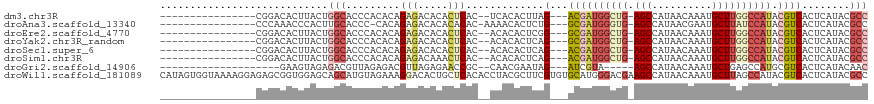
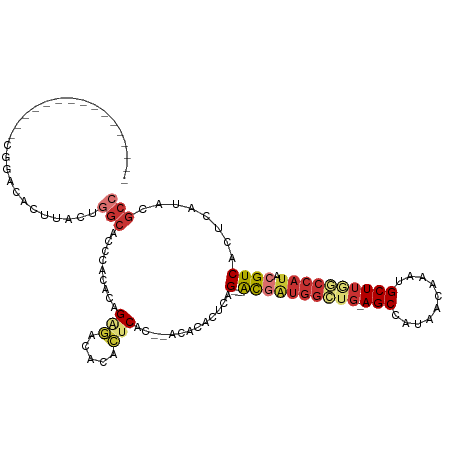
>dm3.chr3R 3452658 96 - 27905053 ----------------CGGACACUUACUGGCACCCACACAGAGACACACUCAC--UCACACUUAG---ACGAUGGCUG-AGCCAUAACAAAUGCUUGGCCAUACGUCACUCAUACGCC ----------------............(((.........(((.....)))..--.........(---((((((((..-(((..........)))..))))).))))........))) ( -26.70, z-score = -4.35, R) >droAna3.scaffold_13340 3223390 96 - 23697760 ----------------CCCAAACCCACUUGCACCC-CACAGAGACACACACAC-AAAACACUCUG---GCGAUGGGUG-AGCCAUAACGAAUGCUUAUCCAUACGUCACUCAUACGCC ----------------.............((....-..(((((..........-......)))))---((((((((((-(((..........)))))))))).))).........)). ( -21.19, z-score = -2.61, R) >droEre2.scaffold_4770 3713406 96 - 17746568 ----------------CGGACACUUACUGGCACCCACACAGAGACACACUCAC--ACACACUCGG---GCGAUGGCUG-AGCCAUAACAAAUGCUUGGCCAUACGUCACUCAUACGCC ----------------............(((.........(((.....)))..--.........(---((((((((..-(((..........)))..))))).))))........))) ( -25.90, z-score = -2.82, R) >droYak2.chr3R_random 1074180 96 - 3277457 ----------------CGGACACUUACUGGCACCCACACAGAGACACACUCAC--ACACACUCAG---GCGAUGGCUG-AGCCAUAACAAAUGCUUGGCCAUACGUCACUCAUACGCC ----------------............(((.........(((.....)))..--.........(---((((((((..-(((..........)))..))))).))))........))) ( -26.10, z-score = -3.21, R) >droSec1.super_6 3545252 96 - 4358794 ----------------CGGACACUUACUGGCACCCACACAGAGACACACUCAC--ACACACUCAG---ACGAUGGCUG-AGCCAUAACAAAUGCUUGGCCAUACGUCACUCAUACGCC ----------------............(((.........(((.....)))..--.........(---((((((((..-(((..........)))..))))).))))........))) ( -26.70, z-score = -4.26, R) >droSim1.chr3R 3505559 96 - 27517382 ----------------CGGACACUUACUGGCACCCACACAGAGACAAACUCAC--ACACACUCAG---ACGAUGGCUG-AGCCAUAACAAAUGCUUGGCCAUACGUCACUCAUACGCC ----------------............(((.........(((.....)))..--.........(---((((((((..-(((..........)))..))))).))))........))) ( -26.70, z-score = -4.32, R) >droGri2.scaffold_14906 6449225 88 + 14172833 --------------------GAAGUAGAGACGUUAGAGACGUUAGAGAACCGC--CAACGAAUAG---AUCGUA-----AGCCAUAACAAAUGCUGAGCCAUGCGUCACUCAUACAAC --------------------...(((..((((((...)))))).(((....((--..((((....---.)))).-----.))........((((........))))..))).)))... ( -15.30, z-score = -0.89, R) >droWil1.scaffold_181089 5017039 118 + 12369635 CAUAGUGGUAAAAGGAGAGCGGUGGAGCAGCAUGUAGAAAGGACACUGCUCACACCUACGCUUCGUGUGCAUGGGACGAAGCCAUAACAAAUGCUUAGCCAUACGUCACUCAUACGCC ...(((((.....((.((((((((((((((..(((.......))))))))).))))...(((((((.(.....).)))))))..........))))..)).....)))))........ ( -35.30, z-score = -1.05, R) >consensus ________________CGGACACUUACUGGCACCCACACAGAGACACACUCAC__ACACACUCAG___ACGAUGGCUG_AGCCAUAACAAAUGCUUGGCCAUACGUCACUCAUACGCC ............................(((.........(((.....))).................((((((((((.(((..........)))))))))).))).........))) ( -8.94 = -10.24 + 1.30)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:56:34 2011