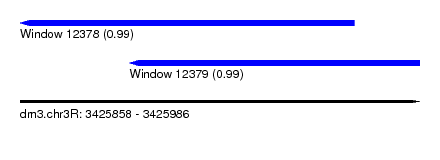
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 3,425,858 – 3,425,986 |
| Length | 128 |
| Max. P | 0.990178 |

| Location | 3,425,858 – 3,425,965 |
|---|---|
| Length | 107 |
| Sequences | 9 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 70.66 |
| Shannon entropy | 0.60094 |
| G+C content | 0.47335 |
| Mean single sequence MFE | -30.34 |
| Consensus MFE | -11.20 |
| Energy contribution | -11.10 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.41 |
| SVM RNA-class probability | 0.990178 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
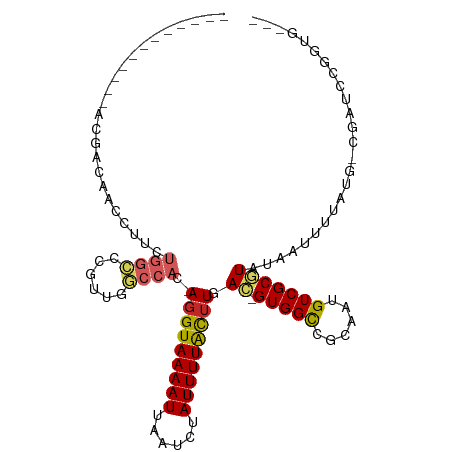
>dm3.chr3R 3425858 107 - 27905053 GGCCAC-AGGUAAAAUUAAUCUAUUUUACUUGAC-GUGGCCGCAAUGUCGCGUAUAAUUUUAUG--CGAUCCGGUGCCCUUCGUUCAUCACCUGCCCAUUCCAAUGAGGUG----- ((((((-(((((((((......)))))))))...-))))))(((..((((((((......))))--))))....)))...........(((((...(((....))))))))----- ( -34.40, z-score = -3.52, R) >droWil1.scaffold_181089 5047395 98 + 12369635 GGCCACAAGGUAAAAUUAAUCUAUUUUAUUUUAU-GUGGCCGCAAUGUCGCAUAUAAUUUUAUGU-CGAACUCCCAUCGCCCA--UACUCUCUAGUGCGGUG-------------- ((((((((((((((((......)))))))))..)-))))))......(((((((......)))).-))).....((((((...--.(((....)))))))))-------------- ( -28.10, z-score = -3.91, R) >droPer1.super_6 2136786 110 - 6141320 GCCCAC-AGGUAAAAUUAAUCUAUUUUGCUUCACGGUGGCCCCAAUGUCGCGUAUAAUUUUAUGGCUGCCACUCUGCCCCUCC-----CCCCCCCGGUGGCAGCUUCGGCUGGACA ((((..-(((((((((......)))))))))....).))).(((..((((((((......))))(((((((((..........-----.......)))))))))..)))))))... ( -31.83, z-score = -0.51, R) >dp4.chr2 26829078 115 - 30794189 GCCCAC-AGGUAAAAUUAAUCUAUUUUGCUUCACGGUGGCCGCAAUGUCGCGUAUAAUUUUAUGGCUGCCACUCUGCCCCUCCCCCCUCCCCCCCGGUGGCAGCUUCGGCUGGACA ..((((-(((((((((......)))))))))...((..(((((......))(((......))))))..))..........................))))((((....)))).... ( -32.90, z-score = -0.47, R) >droYak2.chr3R 21929608 105 + 28832112 GGCCAC-AGGUAAAAUUAAUCUAUUUUACUUGAC-GUGGCCGCAAUGUCGCGUAUAAUUUUAUG--CGAUCCGGUGCCCUUCAUUCAUCACCUGCCCAUUCCAUUAAAG------- ((((((-(((((((((......)))))))))...-))))))(((..((((((((......))))--))))..((((............)))))))..............------- ( -32.20, z-score = -4.51, R) >droAna3.scaffold_13340 3197048 104 - 23697760 GGCCAC-AGGUAAAAUUAAUCUAUUUUACUUGAC-GUGGCCGCAAUGUCGCGUAUAAUUUUAUG--CGAUCCGCUGGUCUCCUCUCAUGAUUCA-UCAUUACACCGAGA------- ((((((-(((((((((......)))))))))...-))))))((...((((((((......))))--))))..))...((((.....((((....-))))......))))------- ( -32.10, z-score = -3.71, R) >droSec1.super_6 3517808 107 - 4358794 GGCCAC-AGGUAAAAUUAAUCUAUUUUACGUGAC-GUGGCCGCAAUGUCGCGUAUAAUUUUAUG--CGAUCCGGUGCCCUUCGUUCAUCCCCUGCCCAUUCCAUUAAGGUG----- ((((((-(.(((((((......))))))).)...-))))))(((..((((((((......))))--))))....))).............(((.............)))..----- ( -29.62, z-score = -2.10, R) >droEre2.scaffold_4770 3687502 107 - 17746568 GGCCAC-AGGUAAAAUUAAUCUAUUUUACUUGAC-GUGGCCGCAAUGUCGCGUAUAAUUUUAUG--CGAUCCGGUGCCCUUCGUUCAUCACCUGCCCAUUCCAUUAAAAUC----- ((((((-(((((((((......)))))))))...-))))))(((..((((((((......))))--))))..((((............)))))))................----- ( -32.20, z-score = -4.20, R) >droGri2.scaffold_14906 7754812 92 - 14172833 GCCCAC-AGGUAAAAUUAAUCUAUUUUGCUUGAC-GUGGCCGCAAUGUCGCAUAUCAUUUUCGGUAUAUGCCCCCAGGUCCCA-CUACCAUCUCC--------------------- .....(-(((((((((......))))))))))..-(((((......)))))...........((((...(((....)))....-.))))......--------------------- ( -19.70, z-score = -1.17, R) >consensus GGCCAC_AGGUAAAAUUAAUCUAUUUUACUUGAC_GUGGCCGCAAUGUCGCGUAUAAUUUUAUG__CGAUCCGCUGCCCUUCAUUCAUCACCUGCCCAUUCCACUAAGG_______ (.((((.(((((((((......)))))))))....)))).)((......))................................................................. (-11.20 = -11.10 + -0.10)

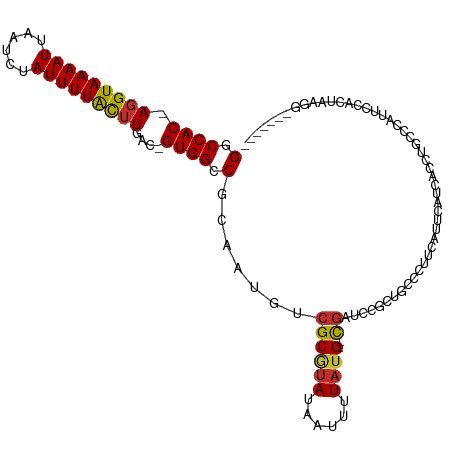
| Location | 3,425,893 – 3,425,986 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 74.40 |
| Shannon entropy | 0.51990 |
| G+C content | 0.45984 |
| Mean single sequence MFE | -29.69 |
| Consensus MFE | -11.84 |
| Energy contribution | -12.08 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.24 |
| SVM RNA-class probability | 0.986588 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 3425893 93 - 27905053 ------------ACGACAACCUUCUGGCCCGUUGGCCAC-AGGUAAAAUUAAUCUAUUUUACUUGAC-GUGGCCGCAAUGUCGCGUAUAAUUUUAUG-CGAUCCGGUG--- ------------(((.........(((((....)))))(-(((((((((......)))))))))).)-)).((((....((((((((......))))-)))).)))).--- ( -32.50, z-score = -3.79, R) >droWil1.scaffold_181089 5047423 103 + 12369635 AGGUUGUGGAUGACAACAACCUUCUGCCCCGAUGGCCACAAGGUAAAAUUAAUCUAUUUUAUUUUAU-GUGGCCGCAAUGUCGCAUAUAAUUUUAUGUCGAACU------- (((((((........)))))))........(.(((((((((((((((((......)))))))))..)-))))))))....(((((((......)))).)))...------- ( -31.00, z-score = -3.25, R) >droPer1.super_6 2136821 90 - 6141320 --------------GAGA----GGGAGGACAUUGCCCAC-AGGUAAAAUUAAUCUAUUUUGCUUCACGGUGGCCCCAAUGUCGCGUAUAAUUUUAUGGCUGCCACUCUG-- --------------(((.----((.(((((((((.((((-(((((((((......)))))))))....))))...))))))).((((......)))).)).)).)))..-- ( -28.60, z-score = -2.22, R) >dp4.chr2 26829118 96 - 30794189 ------------GGGAGAUGACGAGAGGACAUUGCCCAC-AGGUAAAAUUAAUCUAUUUUGCUUCACGGUGGCCGCAAUGUCGCGUAUAAUUUUAUGGCUGCCACUCUG-- ------------(((..(((.(....)..)))..))).(-(((((((((......))))))).....((..(((((......))(((......))))))..))...)))-- ( -28.70, z-score = -1.72, R) >droYak2.chr3R 21929641 93 + 28832112 ------------ACGACAACCUUCUGGCCCGUUGGCCAC-AGGUAAAAUUAAUCUAUUUUACUUGAC-GUGGCCGCAAUGUCGCGUAUAAUUUUAUG-CGAUCCGGUG--- ------------(((.........(((((....)))))(-(((((((((......)))))))))).)-)).((((....((((((((......))))-)))).)))).--- ( -32.50, z-score = -3.79, R) >droAna3.scaffold_13340 3197080 93 - 23697760 ------------ACGACAACCUUCUGGCCCGUUGGCCAC-AGGUAAAAUUAAUCUAUUUUACUUGAC-GUGGCCGCAAUGUCGCGUAUAAUUUUAUG-CGAUCCGCUG--- ------------..(((..((....))...)))((((((-(((((((((......)))))))))...-))))))((...((((((((......))))-))))..))..--- ( -30.40, z-score = -3.59, R) >droSec1.super_6 3517843 93 - 4358794 ------------ACGACAACCUUCUGGCCCGUUGGCCAC-AGGUAAAAUUAAUCUAUUUUACGUGAC-GUGGCCGCAAUGUCGCGUAUAAUUUUAUG-CGAUCCGGUG--- ------------(((.........(((((....)))))(-(.(((((((......))))))).)).)-)).((((....((((((((......))))-)))).)))).--- ( -30.80, z-score = -2.68, R) >droEre2.scaffold_4770 3687537 93 - 17746568 ------------ACGACAACCUUCUGGCCCGUUGGCCAC-AGGUAAAAUUAAUCUAUUUUACUUGAC-GUGGCCGCAAUGUCGCGUAUAAUUUUAUG-CGAUCCGGUG--- ------------(((.........(((((....)))))(-(((((((((......)))))))))).)-)).((((....((((((((......))))-)))).)))).--- ( -32.50, z-score = -3.79, R) >droMoj3.scaffold_6540 27461945 87 + 34148556 -----GCGACUCCUGCCAGACUUCUGCAACGUUGCCCAC-AGGUAAAAUUAAUCUAUUUUGCUUGAC-GUGGUCGCACUGUCGCAUAUCGUUUU----------------- -----(((((...(((..((((.(.(((....)))...(-(((((((((......))))))))))..-).)))))))..)))))..........----------------- ( -25.10, z-score = -2.62, R) >droGri2.scaffold_14906 7754828 104 - 14172833 -----ACAACGACGACUAGACUUCUGGCCCGAUGCCCAC-AGGUAAAAUUAAUCUAUUUUGCUUGAC-GUGGCCGCAAUGUCGCAUAUCAUUUUCGGUAUAUGCCCCCAGG -----...............((...(((...(((((..(-(((((((((......))))))))))..-(((((......)))))...........)))))..)))...)). ( -24.80, z-score = -0.65, R) >consensus ____________ACGACAACCUUCUGGCCCGUUGGCCAC_AGGUAAAAUUAAUCUAUUUUACUUGAC_GUGGCCGCAAUGUCGCGUAUAAUUUUAUG_CGAUCCGGUG___ ........................((((......))))..(((((((((......)))))))))....(((((......)))))........................... (-11.84 = -12.08 + 0.24)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:56:33 2011