
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 3,418,540 – 3,418,631 |
| Length | 91 |
| Max. P | 0.970923 |

| Location | 3,418,540 – 3,418,631 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 69.63 |
| Shannon entropy | 0.61196 |
| G+C content | 0.56430 |
| Mean single sequence MFE | -30.25 |
| Consensus MFE | -14.81 |
| Energy contribution | -16.20 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.958758 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
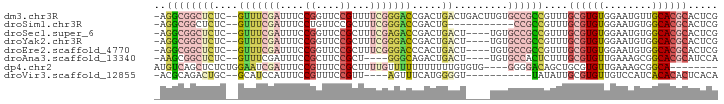
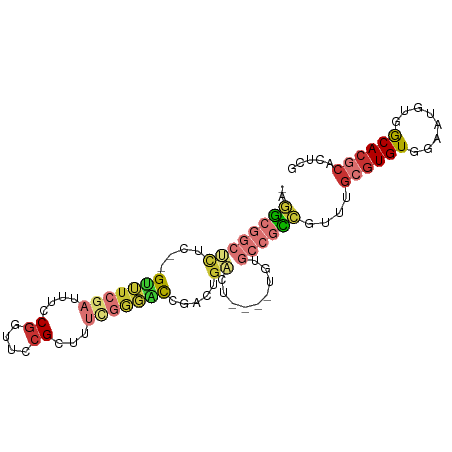
>dm3.chr3R 3418540 91 + 27905053 -AGGCGGCUCUC--GUUUCGAUUUCCGGUUCCGUUUUCGGGACCGACUGACUGACUUGUGCCGCCGUUUGCGUGUGGAAUGUUGCACGCACUCG -.((((((.(..--(((........((((((((....)))))))).......)))..).))))))(..((((((..(....)..))))))..). ( -41.86, z-score = -4.05, R) >droSim1.chr3R 3467175 80 + 27517382 -AGGCGGCUCUC--GUUUCGAUUUCCUGUUCCGCUUUCGGGACCGACUG-----------CCGCCGUUUGCGUGUGGAAUGUGGCACGCACUCG -.((((((..((--((..(((...............)))..).)))..)-----------)))))(..(((((((.(....).)))))))..). ( -33.46, z-score = -2.39, R) >droSec1.super_6 3510121 87 + 4358794 -AGGCGGCUCUC--GUUUCGAUUUCCGGUUCCGCUUUCGAGACCGACUGACU----UGUGCCGCCGUUUGCGUGUGGAAUGUGGCACGCACUCG -.((((((....--(((((((....((....))...)))))))..((.....----.))))))))(..(((((((.(....).)))))))..). ( -34.10, z-score = -1.72, R) >droYak2.chr3R 21920087 87 - 28832112 -AGGCGGCUCUC--GUUUCGAUUUCCGGUUCCGCUUUCGGGACCGACUGACU----UGUGCCGCCGUUUGCGUGUGGAAUGUGGCACGCACUCG -.((((((.(..--(((........((((((((....))))))))...))).----.).))))))(..(((((((.(....).)))))))..). ( -38.20, z-score = -2.72, R) >droEre2.scaffold_4770 3678293 87 + 17746568 -AGGCGGCUCUC--GUUUCGAUUUCCGGUUCCGCUUUCGGGACCCACUGACU----UGUGCCGCCGUUUGCGUGUGGAAUGUGGCACGCACUCG -.((((((.(..--(((.........(((((((....)))))))....))).----.).))))))(..(((((((.(....).)))))))..). ( -35.62, z-score = -2.14, R) >droAna3.scaffold_13340 3189577 83 + 23697760 -AAGCGGCUCUC--GUUUCGAUUUCCGCUUCCGCU----GGGCAGACUGACU----UGUGCCACUCUUUGCGUGUUGAAAGCGGCACGCAUCCA -((((((...((--(...)))...)))))).....----.((((.(......----).))))......(((((((((....))))))))).... ( -26.40, z-score = -0.41, R) >dp4.chr2 26820251 82 + 30794189 AUGUCAGCUCUCUGGAAUCGAUUUCCGUUUCCGCUUUUGUUUUUUUUUUUGUGUG----GGGGACAGCUGCGUGUUGAAAGCGGCA-------- (((.(((((.(((((((.....))))...(((((..................)))----))))).))))))))((((....)))).-------- ( -20.77, z-score = -0.25, R) >droVir3.scaffold_12855 6328048 76 + 10161210 -ACGCAGACUGC--GCAUCCAUUUCCGUUUCCGUU----AGUUUCAUGGGGU-----------UAUAUUGCGUGUUGUCCAUCACACACUCACA -.....(((.((--(((.....((((((.......----......)))))).-----------.....)))))...)))............... ( -11.62, z-score = 0.96, R) >consensus _AGGCGGCUCUC__GUUUCGAUUUCCGGUUCCGCUUUCGGGACCGACUGACU____UGUGCCGCCGUUUGCGUGUGGAAUGUGGCACGCACUCG ..((((((......(((((((....((....))...)))))))................))))))....((((((........))))))..... (-14.81 = -16.20 + 1.39)
| Location | 3,418,540 – 3,418,631 |
|---|---|
| Length | 91 |
| Sequences | 8 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 69.63 |
| Shannon entropy | 0.61196 |
| G+C content | 0.56430 |
| Mean single sequence MFE | -27.88 |
| Consensus MFE | -13.15 |
| Energy contribution | -14.14 |
| Covariance contribution | 0.98 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.84 |
| SVM RNA-class probability | 0.970923 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
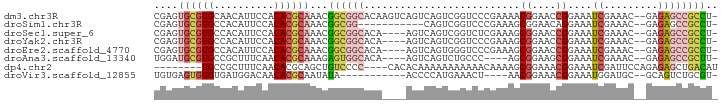
>dm3.chr3R 3418540 91 - 27905053 CGAGUGCGUGCAACAUUCCACACGCAAACGGCGGCACAAGUCAGUCAGUCGGUCCCGAAAACGGAACCGGAAAUCGAAAC--GAGAGCCGCCU- .(..((((((..........))))))..)((((((.(..((..(....(((((.(((....))).)))))....)...))--..).)))))).- ( -33.10, z-score = -2.50, R) >droSim1.chr3R 3467175 80 - 27517382 CGAGUGCGUGCCACAUUCCACACGCAAACGGCGG-----------CAGUCGGUCCCGAAAGCGGAACAGGAAAUCGAAAC--GAGAGCCGCCU- .(..((((((..........))))))..)(((((-----------(..(((.((((....).)))....(....)....)--))..)))))).- ( -29.40, z-score = -1.91, R) >droSec1.super_6 3510121 87 - 4358794 CGAGUGCGUGCCACAUUCCACACGCAAACGGCGGCACA----AGUCAGUCGGUCUCGAAAGCGGAACCGGAAAUCGAAAC--GAGAGCCGCCU- .(..((((((..........))))))..)((((((...----......(((((..((....))..)))))...(((...)--))..)))))).- ( -31.30, z-score = -1.76, R) >droYak2.chr3R 21920087 87 + 28832112 CGAGUGCGUGCCACAUUCCACACGCAAACGGCGGCACA----AGUCAGUCGGUCCCGAAAGCGGAACCGGAAAUCGAAAC--GAGAGCCGCCU- .(..((((((..........))))))..)((((((...----......((((((((....).)).)))))...(((...)--))..)))))).- ( -33.10, z-score = -2.40, R) >droEre2.scaffold_4770 3678293 87 - 17746568 CGAGUGCGUGCCACAUUCCACACGCAAACGGCGGCACA----AGUCAGUGGGUCCCGAAAGCGGAACCGGAAAUCGAAAC--GAGAGCCGCCU- .(..((((((..........))))))..)((((((...----......(((.((((....).))).)))....(((...)--))..)))))).- ( -31.10, z-score = -1.50, R) >droAna3.scaffold_13340 3189577 83 - 23697760 UGGAUGCGUGCCGCUUUCAACACGCAAAGAGUGGCACA----AGUCAGUCUGCCC----AGCGGAAGCGGAAAUCGAAAC--GAGAGCCGCUU- ..(((..((((((((((..........)))))))))).----.)))..(((((..----.)))))(((((...(((...)--))...))))).- ( -30.60, z-score = -1.83, R) >dp4.chr2 26820251 82 - 30794189 --------UGCCGCUUUCAACACGCAGCUGUCCCC----CACACAAAAAAAAAAACAAAAGCGGAAACGGAAAUCGAUUCCAGAGAGCUGACAU --------...............((((((.((...----.......................(....)((((.....)))).)).))))).).. ( -15.60, z-score = -1.35, R) >droVir3.scaffold_12855 6328048 76 - 10161210 UGUGAGUGUGUGAUGGACAACACGCAAUAUA-----------ACCCCAUGAAACU----AACGGAAACGGAAAUGGAUGC--GCAGUCUGCGU- .....((((((........))))))......-----------...((((......----..((....))...))))..((--((.....))))- ( -18.80, z-score = -0.98, R) >consensus CGAGUGCGUGCCACAUUCCACACGCAAACGGCGGCACA____AGUCAGUCGGUCCCGAAAGCGGAACCGGAAAUCGAAAC__GAGAGCCGCCU_ ....((((((..........))))))...((((((..........................((....))....((.........)))))))).. (-13.15 = -14.14 + 0.98)
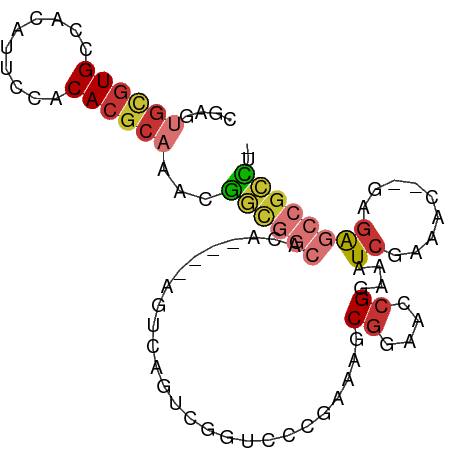
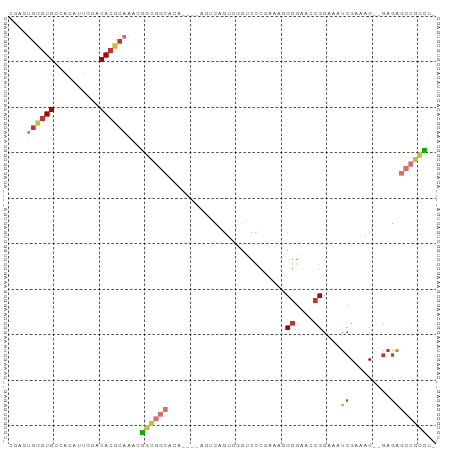
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:56:30 2011