
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 3,415,712 – 3,415,840 |
| Length | 128 |
| Max. P | 0.843614 |

| Location | 3,415,712 – 3,415,840 |
|---|---|
| Length | 128 |
| Sequences | 10 |
| Columns | 152 |
| Reading direction | reverse |
| Mean pairwise identity | 68.19 |
| Shannon entropy | 0.60157 |
| G+C content | 0.46080 |
| Mean single sequence MFE | -32.16 |
| Consensus MFE | -16.62 |
| Energy contribution | -16.18 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.88 |
| SVM RNA-class probability | 0.843614 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
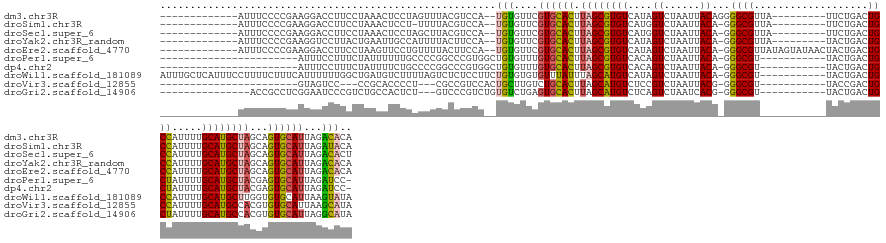
>dm3.chr3R 3415712 128 - 27905053 -------------AUUUCCCCGAAGGACCUUCCUAAACUCCUAGUUUACGUCCA--UGUGUUCGUGCACUUAGCGUGUCAUAGUCUAAUUACAGGGGCGUUA---------UUCUGACUGCCAUUUUGCAUGCUAGCAGUGCAUUAGACACA -------------...........((((.....(((((.....))))).)))).--((((((.(((((((((((((((.....(((......)))((((((.---------....))).))).....))))))))..)))))))..)))))) ( -35.50, z-score = -1.96, R) >droSim1.chr3R 3464341 126 - 27517382 -------------AUUUCCCCGAAGGACCUUCCUAAACUCCU-UUUUACGUCCA--UGUGUUCGUGCACUUAGCGUGUCAUGGUCUAAUUACA-GGGCGUUA---------UUCUGACUGCCAUUUUGCAUGCUAGCAGUGCAUUAGAUACA -------------........((((((...........))))-)).........--((((((.(((((((((((((((.(((((.......((-(((.....---------)))))...)))))...))))))))..)))))))..)))))) ( -31.40, z-score = -1.22, R) >droSec1.super_6 3507299 127 - 4358794 -------------AUUUCCCCGAAGGACCUUCCUAAACUCCUAGCUUACGUCCA--UGUGUUCGUGCACUUAGCGUGUCAUGGUCUAAUUACA-GGGCGUUA---------UUCUGACUGCCAUUUUGCAUGCUAGCAGUGCAUUAGACACU -------------...........((((....(((......))).....)))).--.(((((.(((((((((((((((.(((((.......((-(((.....---------)))))...)))))...))))))))..)))))))..))))). ( -34.50, z-score = -1.48, R) >droYak2.chr3R_random 1055456 127 - 3277457 -------------AUUUCCCCGAAGGUCCUUACUGAAUUGCCAUUUUACUUCCA--UGUGUUCGUGCACUUAGCGUGUCAUAGUCUAAUUACA-GGGCGUUA---------UACUGACUGCCAUUUUGCAUGCUAGCAGUGCAUUAGACACA -------------........((((.......................))))..--((((((.(((((((((((((((....((......)).-.((((((.---------....))).))).....))))))))..)))))))..)))))) ( -30.20, z-score = -0.55, R) >droEre2.scaffold_4770 3675498 136 - 17746568 -------------AUUUCCCCGAAGGACCUUCCUAAGUUCCUGUUUUACUUCCA--UGUGUUCGUGCACUUAGCGUGUCAUAGUCUAAUUACA-GGGCGUUAUAGUAUAACUACUGACUGCCAUUUUGCAUGCUAGCAGUGCAUUAGACACA -------------..........((((.((.....)).))))............--((((((.(((((((((((((((....((......)).-.((((((((((.....))).)))).))).....))))))))..)))))))..)))))) ( -35.80, z-score = -1.92, R) >droPer1.super_6 2123365 116 - 6141320 -----------------------AUUUCCUUUCUAUUUUUUGCCCCGGCCCGUGGCUGUGUUUGUGCACUUAGCGUGUCACAGUCUAAUUACA-GGGCGU-----------UACUGACUGCUAUUUUGCAUGCUACGAGUGCAUUAGAUCC- -----------------------........((((..........(((((...))))).....(((((((((((((((..(((((.....((.-....))-----------....))))).......)))))))..))))))))))))...- ( -29.80, z-score = -0.74, R) >dp4.chr2 26816638 116 - 30794189 -----------------------AUUUCCUUUCUAUUUUCUGCCCCGGCCCGUGGCUGUGUUUGUGCACUUAGCGUGUCACAGUCUAAUUACA-GGGCGU-----------UACUGACUGCUAUUUUGCAUGCUACGAGUGCAUUAGAUCC- -----------------------........((((..........(((((...))))).....(((((((((((((((..(((((.....((.-....))-----------....))))).......)))))))..))))))))))))...- ( -29.80, z-score = -0.61, R) >droWil1.scaffold_181089 7309238 140 - 12369635 AUUUGCUCAUUUCCUUUUCUUUCAUUUUUUGGCUGAUGUCUUUUAGUCUCUCCUUCUGUGUGUGUUUAUUUAGCAUGUCAUAGUCUAAUUACA-GGGCGU-----------UACUGACUGCCAUUUUGCAUGCUUGGUGUGCAUUAAGUAUA ....((((......................((((((......)))))).......(((((((((((.....)))))).)))))..........-))))..-----------((((...(((((((..(....)..)))).)))...)))).. ( -27.90, z-score = -0.87, R) >droVir3.scaffold_12855 6324756 111 - 10161210 -----------------------GUAGUCC---CCGCACCCCU---CGCCGUCCACUGCUUGUCUGCACUUAGCAUGUCUCCGUCUAAUUACG-GGGCGU-----------UACCGACUGCCAUUUUGCAUGCCACGUGUGCAUUAAGCAUA -----------------------.......---..((......---.)).......((((((..(((((...((((((..((((......)))-)((((.-----------.......)))).....)))))).....))))).)))))).. ( -28.60, z-score = -0.57, R) >droGri2.scaffold_14906 6405799 122 + 14172833 ---------------ACCGCCUCGGAAUCCCGUCUGCCACUCU---GUCCCGUCUGUGUCUGAGUGCACUUAGCAUGUCUCAGUCUAAUCACG-GGGCGU-----------UACUGACUGCUAUUUUGCAUGCCACGUGUGCAUUAGGCAUA ---------------...(((((((....)))..((((((...---(((((((....(.((((((((.....)))...))))).).....)))-))))..-----------.......(((......)))......))).)))..))))... ( -38.10, z-score = -1.19, R) >consensus _____________AUUUCCCCGAAGGACCUUCCUAAACUCCUAUUUUACCUCCA__UGUGUUCGUGCACUUAGCGUGUCAUAGUCUAAUUACA_GGGCGU___________UACUGACUGCCAUUUUGCAUGCUAGCAGUGCAUUAGACACA ........................................................(((....((((((.((((((((....((......))...((((...................)))).....))))))))...))))))...))).. (-16.62 = -16.18 + -0.44)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:56:29 2011