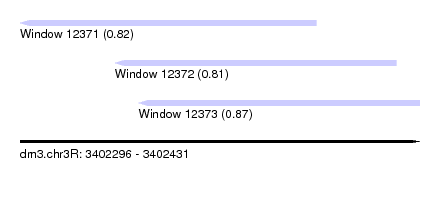
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 3,402,296 – 3,402,431 |
| Length | 135 |
| Max. P | 0.869475 |

| Location | 3,402,296 – 3,402,396 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 62.45 |
| Shannon entropy | 0.63757 |
| G+C content | 0.46831 |
| Mean single sequence MFE | -27.68 |
| Consensus MFE | -12.86 |
| Energy contribution | -13.50 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.819253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 3402296 100 - 27905053 --UUUCUUAAUUCGUGGUAGACCGCACAGGUGGCC-----CUAACUUAUCGGAGCAACCUGCUCCUAGCAAUGGGUUAUCGGAUGGGGAAUCUGACACCUCUCGCAC-------- --.((((..((((((((....))))...(((((((-----(...((....(((((.....))))).))....))))))))))))..)))).................-------- ( -31.40, z-score = -1.32, R) >droSim1.chr3R 3450785 100 - 27517382 --UUUCUUAAUUCGUGGGAGACCACUCAGGUGGUC-----CUAACUUAUUGGAGCAACCUGCUCCUAGCAAUGGGUUAUCAAAUGGGAAAUCUGACACCUCUCGCAC-------- --...........(((((((.....((((((..((-----(((((((((((((((.....)))))....)))))))))......)))..))))))...)))))))..-------- ( -32.20, z-score = -2.64, R) >droSec1.super_6 3494190 100 - 4358794 --UUUCUUAAUUCGUGGGAGACCACUCAGGUGGUC-----CUAACUUAUUGGAGCAACCUGCUCCUAGCAAUGGGUUAUCAAAUGGGGAAUCUGACACCUCUCGCAC-------- --...........(((((((.....((((((..((-----(((((((((((((((.....)))))....)))))))))......)))..))))))...)))))))..-------- ( -31.70, z-score = -2.07, R) >droYak2.chr3R 21902494 87 + 28832112 --UUUCUUAAUUCGUGGGAGACCACU-GGGCUUCC-----CCAACUUAUCAGAAGAAUCUCCUCCUUCCAAUGAGUUAUCAAAUGUGGAACCUGA-------------------- --...........((((....)))).-(((.((((-----.((((((((..((((.........))))..)))))).......)).)))))))..-------------------- ( -22.41, z-score = -1.08, R) >droAna3.scaffold_13340 3174247 115 - 23697760 ACUUUCUGGGUUCAUAAAAAUCUUCCCCCAUCAUCUUAGUCUGAUUUAACUUAUCAGAGGGCUCCCAUCUGUGGCUCAGUGCUAACAGGGCCAGUUACACCUUACAUUUGUGCAU ......((((................)))).........((((((.......))))))((....)).....((((((.((....)).))))))...................... ( -20.69, z-score = 1.22, R) >consensus __UUUCUUAAUUCGUGGGAGACCACUCAGGUGGUC_____CUAACUUAUCGGAGCAACCUGCUCCUAGCAAUGGGUUAUCAAAUGGGGAAUCUGACACCUCUCGCAC________ ...(((((.....((((....))))................((((((((.(((((.....))))).....)))))))).......)))))......................... (-12.86 = -13.50 + 0.64)

| Location | 3,402,328 – 3,402,423 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 75.34 |
| Shannon entropy | 0.45281 |
| G+C content | 0.49540 |
| Mean single sequence MFE | -27.58 |
| Consensus MFE | -14.77 |
| Energy contribution | -15.32 |
| Covariance contribution | 0.55 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.811926 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 3402328 95 - 27905053 GAGCACUUGACCAGCACCCUUGGUUUAUUUCUUAAUUCGUGGUAGACCGCACAGGUGGCCCUAACUUAUCGGAGCAACCUGCUCCUAGCAAUGGG ...(((((((((((.....)))))..............((((....)))).)))))).(((...((....(((((.....))))).))....))) ( -26.90, z-score = -0.76, R) >droSim1.chr3R 3450817 95 - 27517382 GAGCACUUGACCAGGACCCUUGGCUUAUUUCUUAAUUCGUGGGAGACCACUCAGGUGGUCCUAACUUAUUGGAGCAACCUGCUCCUAGCAAUGGG ..((........(((((((((((.(((.....)))...((((....))))))))).))))))........(((((.....)))))..))...... ( -33.10, z-score = -2.25, R) >droSec1.super_6 3494222 95 - 4358794 GAGCACUUGACCAGGACCCUUGGUUUAUUUCUUAAUUCGUGGGAGACCACUCAGGUGGUCCUAACUUAUUGGAGCAACCUGCUCCUAGCAAUGGG ..((........(((((((((((.(((.....)))...((((....))))))))).))))))........(((((.....)))))..))...... ( -33.30, z-score = -2.30, R) >droYak2.chr3R 21902514 78 + 28832112 ----------------CCUUUGGUUUAUUUCUUAAUUCGUGGGAGACCACU-GGGCUUCCCCAACUUAUCAGAAGAAUCUCCUCCUUCCAAUGAG ----------------......................((((....))))(-(((....)))).(((((..((((.........))))..))))) ( -18.10, z-score = -0.67, R) >droEre2.scaffold_4770 3662221 80 - 17746568 GAGAACUUGACCAGUGCCCUUGGUUGACUUCUUAAUUCGUGGGAGACC---------------ACUUAUCGCAGGAACCUCCUCCUUGCAAUGGC (((((.(..(((((.....)))))..).))))).....((((....))---------------)).....((((((......))).)))...... ( -26.50, z-score = -2.58, R) >consensus GAGCACUUGACCAGGACCCUUGGUUUAUUUCUUAAUUCGUGGGAGACCACUCAGGUGGCCCUAACUUAUCGGAGCAACCUGCUCCUAGCAAUGGG ................((((((................((((....))))..(((....)))........(((((.....)))))...))).))) (-14.77 = -15.32 + 0.55)
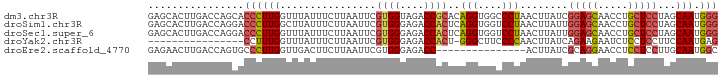

| Location | 3,402,336 – 3,402,431 |
|---|---|
| Length | 95 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 74.50 |
| Shannon entropy | 0.46801 |
| G+C content | 0.49899 |
| Mean single sequence MFE | -26.30 |
| Consensus MFE | -14.89 |
| Energy contribution | -15.64 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.869475 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 3402336 95 - 27905053 GUCCGACAGAGCACUUGACCAGCACCCUUGGUUUAUUUCUUAAUUCGUGGUAGACCGCACAGGUGGCCCUAACUUAUCGGAGCAACCUGCUCCUA .......((.((((((((((((.....)))))..............((((....)))).))))).)).))........(((((.....))))).. ( -25.20, z-score = -0.85, R) >droSim1.chr3R 3450825 95 - 27517382 GUCCGACAGAGCACUUGACCAGGACCCUUGGCUUAUUUCUUAAUUCGUGGGAGACCACUCAGGUGGUCCUAACUUAUUGGAGCAACCUGCUCCUA ........((.((((((((((((...)))))...............((((....))))))))))).))..........(((((.....))))).. ( -32.20, z-score = -2.33, R) >droSec1.super_6 3494230 95 - 4358794 GUCCGACAGAGCACUUGACCAGGACCCUUGGUUUAUUUCUUAAUUCGUGGGAGACCACUCAGGUGGUCCUAACUUAUUGGAGCAACCUGCUCCUA ........((.((((((((((((...)))))...............((((....))))))))))).))..........(((((.....))))).. ( -32.40, z-score = -2.41, R) >droYak2.chr3R 21902522 75 + 28832112 ----------------GUCCG---CCUUUGGUUUAUUUCUUAAUUCGUGGGAGACCACU-GGGCUUCCCCAACUUAUCAGAAGAAUCUCCUCCUU ----------------.....---.....((.....(((((.....((((....))))(-(((....)))).........)))))......)).. ( -17.50, z-score = -0.79, R) >droEre2.scaffold_4770 3662229 80 - 17746568 GUCCGACAGAGAACUUGACCAGUGCCCUUGGUUGACUUCUUAAUUCGUGGGAGACC---------------ACUUAUCGCAGGAACCUCCUCCUU .(((..(.(((((.(..(((((.....)))))..).))))).....((((....))---------------)).....)..)))........... ( -24.20, z-score = -1.78, R) >consensus GUCCGACAGAGCACUUGACCAGGACCCUUGGUUUAUUUCUUAAUUCGUGGGAGACCACUCAGGUGGCCCUAACUUAUCGGAGCAACCUGCUCCUA ................((((((.....)))))).............((((....))))..(((....)))........(((((.....))))).. (-14.89 = -15.64 + 0.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:56:28 2011