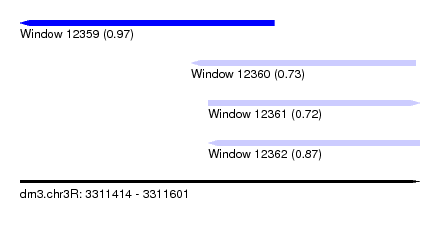
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 3,311,414 – 3,311,601 |
| Length | 187 |
| Max. P | 0.966233 |

| Location | 3,311,414 – 3,311,533 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 86.98 |
| Shannon entropy | 0.20916 |
| G+C content | 0.45475 |
| Mean single sequence MFE | -32.74 |
| Consensus MFE | -28.82 |
| Energy contribution | -28.78 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.76 |
| SVM RNA-class probability | 0.966233 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 3311414 119 - 27905053 AUUUUAAGGGGUUCUGAGGGGGGUUUUAGGGUG-GCUUUCCAGGAGGGGUUAGCGCCUGGAUCCUGUUCAUGUGCACGGGGUGUAAAAAUGGGCGUUAACUCAAUAAAUUAGAAUUUUUU .....((((((((((((((..((((.......)-)))..)).....(((((((((((((.(((((((.(....).))))))).)......))))))))))))......)))))))))))) ( -42.50, z-score = -4.26, R) >droSim1.chr3R 3362951 104 - 27517382 ----------------AGGGGAUUUUUAGGGGGUGCUUUCCAGGAGGGGUUAGCGCCCGGAUCCUGUUCAUGUGCACGGGGUGUAAAAAUGGGCGUUAACUCAAUAAAUUAGAAUUUUUU ----------------.(((((.((((((((((....)))).....(((((((((((((.(((((((.(....).))))))).......)))))))))))))......)))))).))))) ( -32.60, z-score = -1.85, R) >droSec1.super_6 3401098 104 - 4358794 ----------------AGGGGAUUUUUAGGGGGUGCUUUCCAGGAGGGGUUAGCGCCCGGAUCCUGUUCAUGUGCACGGGGUGUAAAAAUGGGCGUUAACUCAAUAAAUUAGAAUUUUUU ----------------.(((((.((((((((((....)))).....(((((((((((((.(((((((.(....).))))))).......)))))))))))))......)))))).))))) ( -32.60, z-score = -1.85, R) >droYak2.chr3R 21806430 97 + 28832112 ----------------------GUUUUUAGGGG-GCCUUCCAGGAGGGGUUAGCGCCUGGAUCCUGUUCAUGUGCACGGGGUGUAAAAAUGGGCGUUAACUCAAUAAAUUAGAAUUUUUU ----------------------..((((((...-..(((....)))(((((((((((((.(((((((.(....).))))))).)......))))))))))))......))))))...... ( -28.10, z-score = -1.00, R) >droEre2.scaffold_4770 3573118 97 - 17746568 ---------------------GUUUUUAGGGGGUG--UUCCAGGAGGGGUUAGCGCCUGGAUCCUGUUCAUGUGCACGGGGUGUAAAAAUGGGCGUUAACUCAAUAAAUUAGAAUUUUUU ---------------------..((((((..((..--..)).....(((((((((((((.(((((((.(....).))))))).)......))))))))))))......))))))...... ( -27.90, z-score = -1.75, R) >consensus ________________AGGGGAUUUUUAGGGGGUGCUUUCCAGGAGGGGUUAGCGCCUGGAUCCUGUUCAUGUGCACGGGGUGUAAAAAUGGGCGUUAACUCAAUAAAUUAGAAUUUUUU ........................(((..((((....))))..)))(((((((((((((.(((((((.(....).))))))).)......)))))))))))).................. (-28.82 = -28.78 + -0.04)


| Location | 3,311,494 – 3,311,599 |
|---|---|
| Length | 105 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 78.16 |
| Shannon entropy | 0.33780 |
| G+C content | 0.56072 |
| Mean single sequence MFE | -29.32 |
| Consensus MFE | -21.04 |
| Energy contribution | -21.44 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.725260 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 3311494 105 - 27905053 CACAGAAUUAGACUGUGUUAUGGUCAUGGCUGUGGCAUGCGGCCGGGCAUGACCACCGGA-----UGGGGGAUUUUAAGGGGUUCUGAGGGGGGUUUUAGGGUGGCUUUC ..(((((((...((..(((.((((((((.((..(((.....))))).))))))))((...-----...)))))....)).)))))))..(..((((.......))))..) ( -31.80, z-score = -0.78, R) >droSim1.chr3R 3363031 89 - 27517382 CACAGAAUUAGACUGUGUUAUGGUCAUGGCUGUGGCAUGCGCACGGGCGUGACCACCGGAGGGGGAGGGGAUUUUUAGGGGGUGCUUUC--------------------- (((((.......)))))...((((((((.((((((....).))))).))))))))((....)).(((((.((((.....)))).)))))--------------------- ( -30.80, z-score = -2.37, R) >droSec1.super_6 3401178 89 - 4358794 CACAGAAUUAGACUGUGUUAUGGUCAUGGCUGUGGCAUGCGCACGGGCGUGACCACCGGAGGAGGAGGGGAUUUUUAGGGGGUGCUUUC--------------------- (((((.......)))))...((((((((.((((((....).))))).)))))))).........(((((.((((.....)))).)))))--------------------- ( -29.70, z-score = -2.03, R) >droYak2.chr3R 21806510 82 + 28832112 UACAGAAUUAGACUGUGUUAUGGUCAUGGCUGCGGCAUGCGCCCGGGCGUGACCAACGGA-----UGGGGGUUUUUAGGGGGCCUUC----------------------- (((((.......)))))...((((((((.(((.(((....)))))).)))))))).....-----.((((((((.....))))))))----------------------- ( -31.80, z-score = -2.08, R) >droEre2.scaffold_4770 3573198 81 - 17746568 UACAGAAUUAGACUGUGUUAUGGUCAUGGCUGUGGCAUGCG-CCGGGCGUGACCAACGGA-----UGGGGGUUUUUAGGGGGUGUUC----------------------- (((....(((((((.((((.((((((((.((..(((....)-)))).))))))))...))-----)).))...)))))...)))...----------------------- ( -22.50, z-score = -0.31, R) >consensus CACAGAAUUAGACUGUGUUAUGGUCAUGGCUGUGGCAUGCGCCCGGGCGUGACCACCGGA_____UGGGGGUUUUUAGGGGGUGCUC_______________________ (((((.......)))))...((((((((.(((.(((....)))))).))))))))....................................................... (-21.04 = -21.44 + 0.40)

| Location | 3,311,502 – 3,311,601 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 79.93 |
| Shannon entropy | 0.30626 |
| G+C content | 0.55770 |
| Mean single sequence MFE | -17.84 |
| Consensus MFE | -12.06 |
| Energy contribution | -12.86 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.718893 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
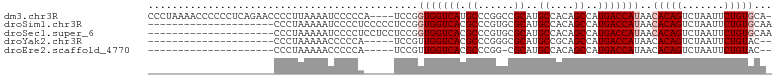
>dm3.chr3R 3311502 99 + 27905053 CCCUAAAACCCCCCUCAGAACCCCUUAAAAUCCCCCA----UCCGGUGGUCAUGCCCGGCCGCAUGCCACAGCCAUGACCAUAACACAGUCUAAUUCUGUGCA- .....................................----....((((((((((..(((.....)))...).)))))))))..(((((.......)))))..- ( -21.50, z-score = -2.45, R) >droSim1.chr3R 3363040 83 + 27517382 ---------------------CCCUAAAAAUCCCCUCCCCCUCCGGUGGUCACGCCCGUGCGCAUGCCACAGCCAUGACCAUAACACAGUCUAAUUCUGUGCAA ---------------------........................(((((((.((..(((.(....)))).))..)))))))..(((((.......)))))... ( -18.40, z-score = -2.17, R) >droSec1.super_6 3401187 83 + 4358794 ---------------------CCCUAAAAAUCCCCUCCUCCUCCGGUGGUCACGCCCGUGCGCAUGCCACAGCCAUGACCAUAACACAGUCUAAUUCUGUGCAA ---------------------........................(((((((.((..(((.(....)))).))..)))))))..(((((.......)))))... ( -18.40, z-score = -2.20, R) >droYak2.chr3R 21806517 76 - 28832112 ---------------------CCCUAAAAACCCCCA-----UCCGUUGGUCACGCCCGGGCGCAUGCCGCAGCCAUGACCAUAACACAGUCUAAUUCUGUAC-- ---------------------...............-----...((((((((.((...(((....)))...))..))))))..))((((.......))))..-- ( -15.60, z-score = -0.46, R) >droEre2.scaffold_4770 3573205 75 + 17746568 ---------------------CCCUAAAAACCCCCA-----UCCGUUGGUCACGCCCGG-CGCAUGCCACAGCCAUGACCAUAACACAGUCUAAUUCUGUAC-- ---------------------...............-----...((((((((.((..((-(....)))...))..))))))..))((((.......))))..-- ( -15.30, z-score = -1.68, R) >consensus _____________________CCCUAAAAAUCCCCA_____UCCGGUGGUCACGCCCGGGCGCAUGCCACAGCCAUGACCAUAACACAGUCUAAUUCUGUGCA_ .............................................(((((((.((......))..((....))..)))))))..(((((.......)))))... (-12.06 = -12.86 + 0.80)
| Location | 3,311,502 – 3,311,601 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 79.93 |
| Shannon entropy | 0.30626 |
| G+C content | 0.55770 |
| Mean single sequence MFE | -27.60 |
| Consensus MFE | -21.34 |
| Energy contribution | -21.74 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.874969 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
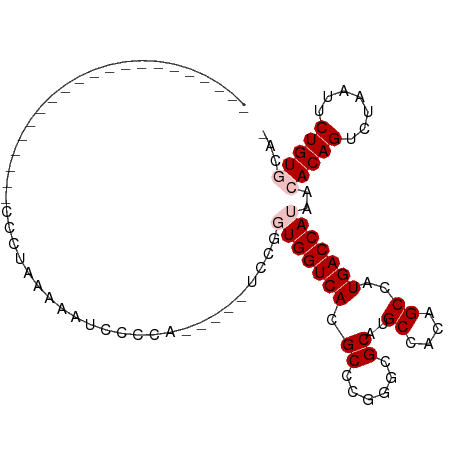
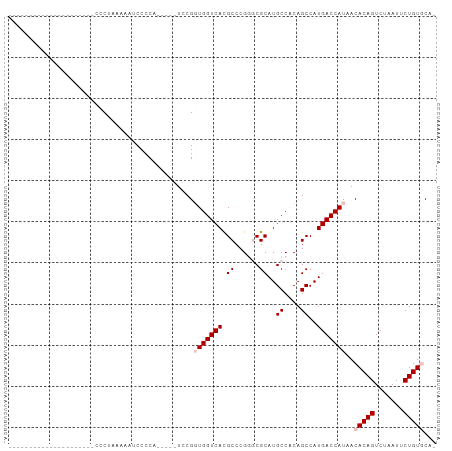
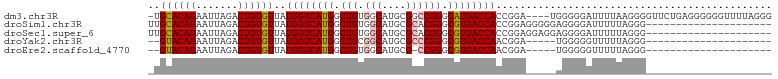
>dm3.chr3R 3311502 99 - 27905053 -UGCACAGAAUUAGACUGUGUUAUGGUCAUGGCUGUGGCAUGCGGCCGGGCAUGACCACCGGA----UGGGGGAUUUUAAGGGGUUCUGAGGGGGGUUUUAGGG -.((((((.......))))))..((((((((.((..(((.....))))).))))))))((...----(..(((((((....)))))))..)..))......... ( -29.40, z-score = -0.56, R) >droSim1.chr3R 3363040 83 - 27517382 UUGCACAGAAUUAGACUGUGUUAUGGUCAUGGCUGUGGCAUGCGCACGGGCGUGACCACCGGAGGGGGAGGGGAUUUUUAGGG--------------------- ..((((((.......))))))..((((((((.((((((....).))))).))))))))((.(((((........))))).)).--------------------- ( -29.60, z-score = -2.83, R) >droSec1.super_6 3401187 83 - 4358794 UUGCACAGAAUUAGACUGUGUUAUGGUCAUGGCUGUGGCAUGCGCACGGGCGUGACCACCGGAGGAGGAGGGGAUUUUUAGGG--------------------- ..((((((.......))))))..((((((((.((((((....).))))).))))))))((.(((((........))))).)).--------------------- ( -30.60, z-score = -3.34, R) >droYak2.chr3R 21806517 76 + 28832112 --GUACAGAAUUAGACUGUGUUAUGGUCAUGGCUGCGGCAUGCGCCCGGGCGUGACCAACGGA-----UGGGGGUUUUUAGGG--------------------- --.............((.((((.((((((((.(((.(((....)))))).))))))))...))-----)).))..........--------------------- ( -26.30, z-score = -1.37, R) >droEre2.scaffold_4770 3573205 75 - 17746568 --GUACAGAAUUAGACUGUGUUAUGGUCAUGGCUGUGGCAUGCG-CCGGGCGUGACCAACGGA-----UGGGGGUUUUUAGGG--------------------- --.............((.((((.((((((((.((..(((....)-)))).))))))))...))-----)).))..........--------------------- ( -22.10, z-score = -0.66, R) >consensus _UGCACAGAAUUAGACUGUGUUAUGGUCAUGGCUGUGGCAUGCGCCCGGGCGUGACCACCGGA_____AGGGGAUUUUUAGGG_____________________ ..((((((.......))))))..((((((((.(((.(((....)))))).)))))))).............................................. (-21.34 = -21.74 + 0.40)
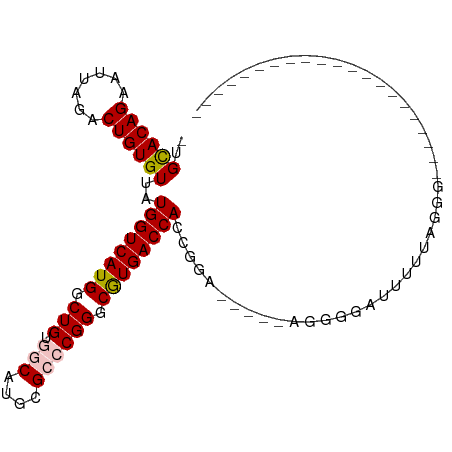
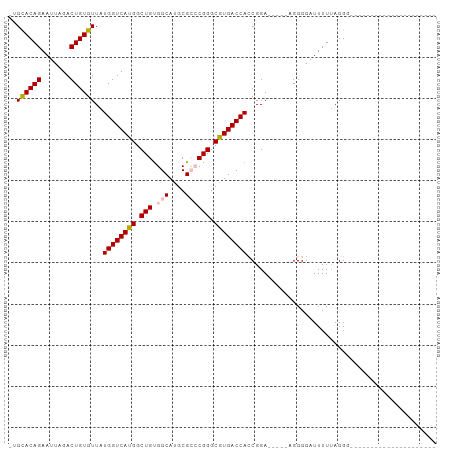
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:56:19 2011