| Sequence ID | dm3.chr3R |
|---|---|
| Location | 3,284,476 – 3,284,589 |
| Length | 113 |
| Max. P | 0.813492 |

| Location | 3,284,476 – 3,284,589 |
|---|---|
| Length | 113 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 87.56 |
| Shannon entropy | 0.25691 |
| G+C content | 0.43289 |
| Mean single sequence MFE | -27.50 |
| Consensus MFE | -15.81 |
| Energy contribution | -15.70 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.813492 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 3284476 113 - 27905053 ------GCUCGUACCGCUUGGUUGCAUGCAAUCUAGAAACGAUUCUAAACAAAAGGAAUCCGCUUUCAUAAUUUCCCCAUGGAACGCGCAUUCUUUUUAUUGCCACUAAGCGACCGAAC ------..(((...((((((((.(((.......(((((....)))))...(((((((((.(((((((((.........)))))).))).)))))))))..))).))))))))..))).. ( -33.00, z-score = -4.10, R) >droSim1.chr3R 3329104 113 - 27517382 ------GCUCGUGCCGCUUCGUUGCAUGCAAUCUAGAAACGAUUCUAAACAAAAGGAAUCCGCUUUCAUAAUUUCCCCAUGGAACGCGCAUUCUUUUUAUUGCCACUAAGCGACCGAAC ------..(((.(.(((((.((.(((.......(((((....)))))...(((((((((.(((((((((.........)))))).))).)))))))))..))).)).))))).)))).. ( -30.20, z-score = -3.05, R) >droSec1.super_6 3375471 113 - 4358794 ------GCUCGUGCCGCUUCGUUGCAUGCAAUCUAGAAACGAUUCUAAACAAAAGGAAUCCGCUUUCAUAAUUUCCCCAUGGAACGCGCAUUCUUUUUAUUGCCACUAAGCGACCGAAC ------..(((.(.(((((.((.(((.......(((((....)))))...(((((((((.(((((((((.........)))))).))).)))))))))..))).)).))))).)))).. ( -30.20, z-score = -3.05, R) >droYak2.chr3R 21770351 113 + 28832112 ------GUUCGUACCGCUUGGUUGCAUGCAAUCUAGAAACGAUUCUAAACAAAAGGAAUCCGCUUUCAUAAUUUCCCCAUGGAACGCGCAUUCUUUUUAUUGCCACUAAGCGACCGAAC ------(((((...((((((((.(((.......(((((....)))))...(((((((((.(((((((((.........)))))).))).)))))))))..))).))))))))..))))) ( -35.70, z-score = -5.01, R) >droEre2.scaffold_4770 3549805 114 - 17746568 -----CGCUCGUACCGCUUGGUCGCAUGCAAUCUAGAAACGAUUCUAAACAAAAGGAAUCCGCUUUCAUAAUUUCCCCAUGGAGCGCGCAUUCUUUUUAUUGCCACUAAGCGACCGAAC -----...(((...((((((((.(((.......(((((....)))))...(((((((((.(((((((((.........)))))).))).)))))))))..))).))))))))..))).. ( -32.70, z-score = -3.16, R) >droAna3.scaffold_13340 20626239 113 + 23697760 ------GGCCGCACCGCUUGGCUGCAUGCAAUCUAGAAACGAUUCUAAACAAAAGGAAUCCGCUUUCAUAAUUUCCCCAUGGAACGCGCAUUCUUUUUAUUGCCACUAAGCGACCGAAC ------...((...(((((((.((...(((((.(((((....)))))...(((((((((.(((((((((.........)))))).))).)))))))))))))))))))))))..))... ( -29.30, z-score = -2.30, R) >dp4.chr2 4101848 117 + 30794189 --GUAUGGGCACGCCGCUUCGCUGCAUGCAAUCUAGAAACGAUUCUAAACAAAAGAAAUCCGCUUUCAUAAUUUCCCUAUGGAAUGAGCAUUCUUUUUAUUGCCACAAAGCGACCGAAC --...(((((.....)).(((((....(((((.(((((....)))))...(((((((....((((.......((((....)))).)))).))))))).))))).....))))))))... ( -22.80, z-score = -0.29, R) >droPer1.super_6 4131288 117 + 6141320 --GUAUGGGCACGCCGCUUCGCUGCAUGCAAUCUAGAAACGAUUCUAAACAAAAGAAAUCCGCUUUCAUAAUUUCCCUAUGGAAUGAGCAUUCUUUUUAUUGCCACUAAGCGACCGAAC --...(((((.....)).(((((....(((((.(((((....)))))...(((((((....((((.......((((....)))).)))).))))))).))))).....))))))))... ( -22.80, z-score = -0.35, R) >droWil1.scaffold_181089 7165653 114 - 12369635 GCUCUUUCACGCACACAUG-GCUGAAUGCAAUCUAGAAACGAUUCUAAACAAAAGAAAUCCGCUUUCAUAAUUUCCCUAUGGAAUGCGCAUUCUUUUUAUUGCCACUAAGCGACC---- .........(((.....((-((.((((((....(((((....)))))..............(((((((((.......))))))).))))))))........))))....)))...---- ( -24.20, z-score = -2.44, R) >droVir3.scaffold_12855 3982852 110 + 10161210 ------ACACGCAGACACAUACUGCAUGCAAUCUAGAAACGAUUCUAAACAAAAGAAAUCCGCUUUCAUAAUUUCCCUAUGGAAUGCACAUUCUUUUUAUUGCCACUAAGCGACCA--- ------....((((.......))))..(((((.(((((....)))))...(((((((....(((((((((.......))))))).))...))))))).))))).............--- ( -19.20, z-score = -2.48, R) >droMoj3.scaffold_6540 27634787 110 + 34148556 ------GCAAGCAGACACAUGCUGCAUGCAAUCUAGAAACGAUUCUAAACAAAAGAAAUCCGCUUUCAUAAUUUCCCUGUGGAAUGCACAUUCUUUUUAUUGCCACUAAGCGACCG--- ------(((.((((.(....))))).)))....(((((....)))))...(((((((....(((((((((.......))))))).))...)))))))..((((......))))...--- ( -22.40, z-score = -1.68, R) >consensus ______GCUCGCACCGCUUCGCUGCAUGCAAUCUAGAAACGAUUCUAAACAAAAGGAAUCCGCUUUCAUAAUUUCCCCAUGGAACGCGCAUUCUUUUUAUUGCCACUAAGCGACCGAAC ....................(((....(((((.(((((....)))))...(((((((...(((((((((.........)))))).)))..))))))).))))).....)))........ (-15.81 = -15.70 + -0.11)

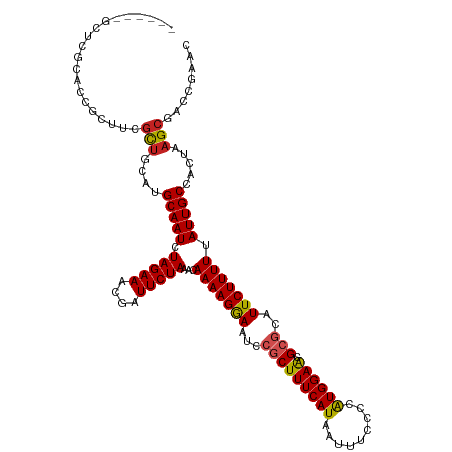
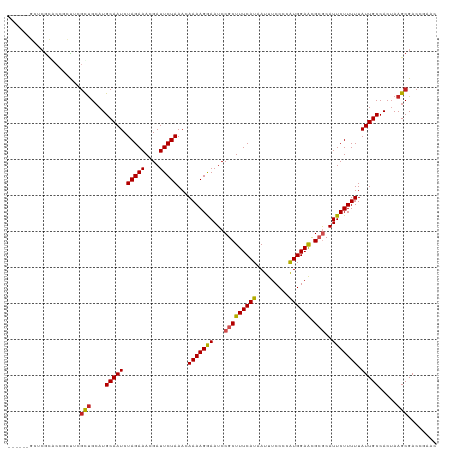
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:56:15 2011