
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 650,423 – 650,524 |
| Length | 101 |
| Max. P | 0.989556 |

| Location | 650,423 – 650,524 |
|---|---|
| Length | 101 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.46 |
| Shannon entropy | 0.18977 |
| G+C content | 0.32884 |
| Mean single sequence MFE | -25.81 |
| Consensus MFE | -21.43 |
| Energy contribution | -21.44 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.88 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.37 |
| SVM RNA-class probability | 0.989556 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
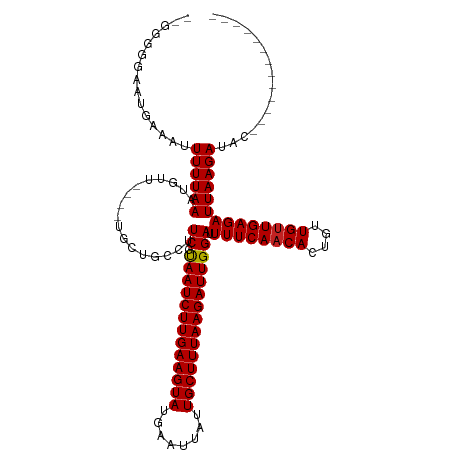
>dm3.chr2L 650423 101 - 23011544 --GGGGGAAUGAAAUUUUUAAAUGUU----UGCUGCCUUCGUAAUCUUGAAGUAUGAAUUAUUGCUUUAAGAUUGGAUUUUCAACACUGUUGUUGAGAUUAAGAUAC------------- --(((((...(((((........)))----).)..))))).(((((((((((((........)))))))))))))...((((((((....)))))))).........------------- ( -25.30, z-score = -3.12, R) >droSec1.super_14 632331 100 - 2068291 ---GGGGAAUGAAAUUUUUAAAUGUU----UGCUGCCUUCGUAAUCUUGAAGUAUGAAUUAUUGCUUUAAGAUUGGAUUUUCAACACUGUUGUUGAGAUUAAGAUAC------------- ---.(((...(((((........)))----).)..)))((.(((((((((((((........))))))))))))))).((((((((....)))))))).........------------- ( -22.10, z-score = -2.17, R) >droYak2.chr2L 641117 101 - 22324452 --GGGGGAAUGAAAUUUUUAAAUGUU----UGCUGCCUUCGUAAUCUUGAAGUAUGAAUUAUUGCUUUAAGAUUGGAUUUUCAACACUGUUGUUGAGAUUAAGAUAC------------- --(((((...(((((........)))----).)..))))).(((((((((((((........)))))))))))))...((((((((....)))))))).........------------- ( -25.30, z-score = -3.12, R) >droEre2.scaffold_4929 708858 101 - 26641161 --GGGGGAAUGAAAUUUUUAAAUGUU----UGCUGCCUUCGUAAUCUUGAAGUAUGAAUUAUUGCUUUAAGAUUGGAUUUUCAACACUGUUGUUGAGAUUAAGAUAC------------- --(((((...(((((........)))----).)..))))).(((((((((((((........)))))))))))))...((((((((....)))))))).........------------- ( -25.30, z-score = -3.12, R) >droAna3.scaffold_12916 1642652 103 - 16180835 GCGGGGGAAUGAAAUUUUUAAAUGUU----UGCUGCCUUCGUAAUCUUGAAGUAUGAAUUAUUGCUUUAAGAUUGGAUUUUCAACACUGUUGUUGAGAUUAAGAUAC------------- (((((((...(((((........)))----).)..)))))))((((((((((((........))))))))))))....((((((((....)))))))).........------------- ( -27.60, z-score = -3.53, R) >dp4.chr4_group3 2618375 101 + 11692001 --GGGAGGAGGAAAUUUUUAAAUGUU----UGCUGCCUUCGUAAUCUUGAAGUAUGAAUUAUUGCUUUAAGAUUGGAUUUUCAACACUGUUGUUGAGAUUAAGAUAC------------- --.(((((((.((((........)))----).)).))))).(((((((((((((........)))))))))))))...((((((((....)))))))).........------------- ( -29.70, z-score = -4.35, R) >droPer1.super_8 3708363 101 + 3966273 --GGGAGGAGGAAAUUUUUAAAUGUU----UGCUGCCUUCGUAAUCUUGAAGUAUGAAUUAUUGCUUUAAGAUUGGAUUUUCAACACUGUUGUUGAGAUUAAGAUAC------------- --.(((((((.((((........)))----).)).))))).(((((((((((((........)))))))))))))...((((((((....)))))))).........------------- ( -29.70, z-score = -4.35, R) >droWil1.scaffold_180772 6326558 94 + 8906247 ---------GAAAAUUUUUAAAUGUU----UGCUGCCUUCGUAAUCUUGAAGUAUGAAUUAUUGCUUUAAGAUUGGAUUUUCAACACUGUUGUUGAGAUUAAGAUAC------------- ---------......((((((.....----........((.(((((((((((((........))))))))))))))).((((((((....))))))))))))))...------------- ( -21.50, z-score = -2.90, R) >droVir3.scaffold_12963 19131781 114 - 20206255 --GGUGGAAUGAAAUUUUUAAAUGUU----UGCUGCCUUCGUAAUCUUGAAGUAUGAAUUAUUGCUUUAAGAUUGGAUUUUCAACACUGUUGUUGAGAUUAAGAUACCCCGGACCAGACG --.....................(((----((...((.((.(((((((((((((........))))))))))))))).((((((((....))))))))............))..))))). ( -27.10, z-score = -2.18, R) >droMoj3.scaffold_6500 8276191 114 + 32352404 --GGGGGAAUGAAAUUUUUAAAUGUU----UGCUGCCUUCGUAAUCUUGAAGUAUGAAUUAUUGCUUUAAGAUUGGAUUUUCAACACUGUUGUUGAGAUUAAGAUACCCCGGCCCAGACC --((((....................----........((.(((((((((((((........))))))))))))))).((((((((....))))))))........)))).......... ( -29.00, z-score = -2.03, R) >droGri2.scaffold_15252 7017541 106 - 17193109 ----UGGGAAUGAAAUUUUAAAUGUUCUGCUUCUGCCUUCGCAAUCUUGAAGUAUGAAUUAUUGCUUUAAGAUUGGAUUUUCAACACUGUUGCUGAGAUUAAGAUACCGC---------- ----.((......(((((((........((....))..((.(((((((((((((........)))))))))))))))................)))))))......))..---------- ( -21.30, z-score = -0.77, R) >consensus __GGGGGAAUGAAAUUUUUAAAUGUU____UGCUGCCUUCGUAAUCUUGAAGUAUGAAUUAUUGCUUUAAGAUUGGAUUUUCAACACUGUUGUUGAGAUUAAGAUAC_____________ ...............((((((.................((.(((((((((((((........))))))))))))))).((((((((....))))))))))))))................ (-21.43 = -21.44 + 0.01)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:06:30 2011