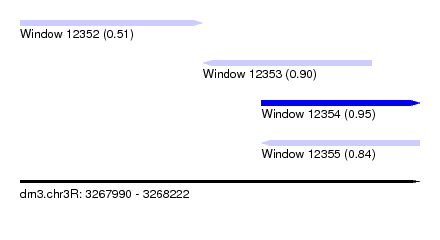
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 3,267,990 – 3,268,222 |
| Length | 232 |
| Max. P | 0.951384 |

| Location | 3,267,990 – 3,268,096 |
|---|---|
| Length | 106 |
| Sequences | 8 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 69.79 |
| Shannon entropy | 0.57530 |
| G+C content | 0.49205 |
| Mean single sequence MFE | -21.86 |
| Consensus MFE | -10.12 |
| Energy contribution | -10.28 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.09 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.508496 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 3267990 106 + 27905053 UGACACUGAAAGCCGCUGUCAGCUGCCAAAUAGCUACAAACGGU-UUAGCCCCC-GGCUAGCACACAAAUACAUUCAUAUACAUAACACGGCACCCGGCACACAAAUC ...........((((.(((((((((.....))))).......((-((((((...-)))))).)).........................))))..))))......... ( -21.20, z-score = -0.72, R) >droSim1.chr3R 3312414 107 + 27517382 UGACACUGAAAGCCGCUGUCAGCUGCCAAAUAGCUGCAAACGGU-UUAGCCCCCUGGCUAACACACAAAUACAUACAUAUACAUAAUACGGCACCCGGCACACAAAUC .....(((...((((.((.((((((.....))))))))...(..-((((((....))))))..)........................))))...))).......... ( -22.50, z-score = -0.79, R) >droSec1.super_6 3359105 107 + 4358794 UGACACUGAAAGCCGCUGUCAGCUGCCAAAUAGCUGCAAACGGU-UUAGCCCCCUGGCUAACACACAAAUACCUACAUAUACAUAAUACGGCACCCGGCACACAAAUC .....(((...((((.((.((((((.....))))))))...(..-((((((....))))))..)........................))))...))).......... ( -22.50, z-score = -0.80, R) >droYak2.chr3R_random 962713 100 + 3277457 UGACACUGAAAGCCGCUGUCAACUGCCAAAUAGCUGCAAACGGC-UGAGCCCCC-GGCUAAUAUGC-----UAUACACAUGCAUAUUACGGCACCCA-UACACAAGUC (((((.((.....)).)))))..((((...((((((....))))-))((((...-))))(((((((-----.........)))))))..))))....-.......... ( -26.30, z-score = -1.74, R) >droEre2.scaffold_4770 3533579 80 + 17746568 UGAAACUGAGAGCCGCUGUCAGCUGCCAAAUAGCUGCAAACGGC-UUAGCCCCC-GGCUAAUAUGC-----UAUACACAUGCAAAUC--------------------- .........((((((.((.((((((.....))))))))..))))-))((((...-))))....(((-----.........)))....--------------------- ( -22.00, z-score = -1.48, R) >droAna3.scaffold_13340 3051501 85 + 23697760 UGACACUGAAAGCCGCUGUCAGCUUGGAAAUUGCUACAAGCCCUUUCAGGCCCUCGGCUAAUAUAC------AUAUUAUGACGCCG----GAACC------------- .....(((((((..(((((.(((.........))))).))).)))))))....(((((..(((...------....)))...))))----)....------------- ( -21.10, z-score = -1.17, R) >dp4.chr2 26670829 95 + 30794189 UGACACUGAAAGCCGCUGUCAGCUGCCAAAUAGCUACAAACGCUCUUAG-CCCCCGGCUAAUAUAC------AUAUUAUGGCGCCCUCUCGAAUCUCGAAUC------ (((((.((.....)).)))))((.((((...(((.......))).((((-((...)))))).....------......))))))....(((.....)))...------ ( -20.50, z-score = -1.18, R) >droPer1.super_6 1988585 89 + 6141320 UGACACUGAAAGCCGCUGUCAGCUGCCAAAUAGCUACAAACGCUCUUAG-CCCCCGGCUAAUAUAC------AUAUUAUGGCGCCCUCUCGAAUCU------------ (((((.((.....)).)))))((.((((...(((.......))).((((-((...)))))).....------......))))))............------------ ( -18.80, z-score = -0.82, R) >consensus UGACACUGAAAGCCGCUGUCAGCUGCCAAAUAGCUACAAACGGU_UUAGCCCCC_GGCUAAUAUAC______AUACAAAUACAUAAUACGGCACCC___ACA______ ..........((((..(((.(((((.....))))))))...(((....)))....))))................................................. (-10.12 = -10.28 + 0.16)
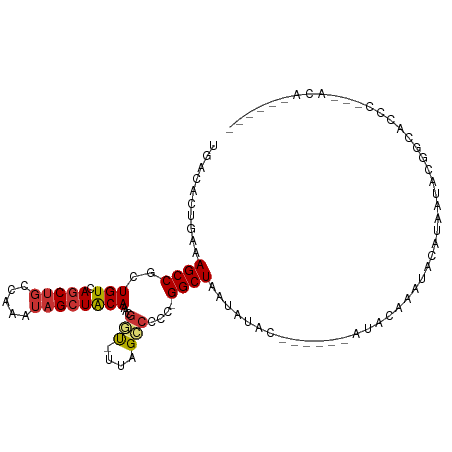
| Location | 3,268,096 – 3,268,194 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 75.00 |
| Shannon entropy | 0.43373 |
| G+C content | 0.47134 |
| Mean single sequence MFE | -30.46 |
| Consensus MFE | -16.84 |
| Energy contribution | -19.40 |
| Covariance contribution | 2.56 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.895257 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 3268096 98 - 27905053 ------AAUUAGCAUUUAGCAUGUUGAGCAAA-UGUCAAGCGAUGGGUUUGGGUUUACGCUUGAAAUUUGGAGCUGAGUUCGGUUUGAGCCUGGUUUUGGGCUUG ------..(((((((.....))))))).((((-(.(((((((.((..(....)..))))))))).)))))((((((....))))))((((((......)))))). ( -30.20, z-score = -2.15, R) >droSim1.chr3R 3312521 98 - 27517382 ------AAUUAGCAUUUAGCAGGUUGAGCGAA-UGUCAAGCGAUGGGUUCGGGUUCACGCUUGAAAUUCGGAACCGAGUUCGGUUUGAGCCUGGUUUUGGGCUUG ------....(((....(.((((((...((((-(.(((((((.((..(....)..))))))))).)))))((((((....)))))).)))))).).....))).. ( -36.80, z-score = -3.34, R) >droSec1.super_6 3359212 98 - 4358794 ------AAUUAGCAUUUAGCAGGUUGAGCGAA-UGUCAAGCGAUGGGUUCGGGUUCACGCUUGAAAUUCGGAACCGAGUUCGGCUUGAGCCAGGUUUUGGGCUUG ------.............(((((((((((((-(.(((((((.((..(....)..))))))))).))))(....)..))))))))))((((........)))).. ( -33.80, z-score = -2.13, R) >droYak2.chr3R_random 962813 98 - 3277457 ------AAUUAGCAUUUAGCAGGUUGAGCGAA-UGUCAAGCGAUGGGUUCGAGUUCACGCUUGAAAUUUGAAGCCGAGUUCGGUUUAAGCCUGGUUUUGGGUUUG ------......((...(.((((((...((((-(.(((((((.((..(....)..))))))))).)))))((((((....)))))).)))))).)..))...... ( -31.80, z-score = -2.57, R) >droWil1.scaffold_181089 7147233 96 - 12369635 GGCAAUAAUUAGCAUUUAGCAGGUUGAACCAGCUGUCAAGAGGCGGAAUUGAAU---------AACAAAAGGGUGGCUCGUGGCAUGAAGCUAGUGAGGGGGGAA ..(....((((((....(((.((.....)).)))((((.(((.((...(((...---------..))).....)).))).)))).....))))))....)..... ( -19.70, z-score = 0.43, R) >consensus ______AAUUAGCAUUUAGCAGGUUGAGCGAA_UGUCAAGCGAUGGGUUCGGGUUCACGCUUGAAAUUCGGAGCCGAGUUCGGUUUGAGCCUGGUUUUGGGCUUG ......(((..((.....))..)))...((((...(((((((.((((......)))))))))))..))))((((((....))))))((((((......)))))). (-16.84 = -19.40 + 2.56)

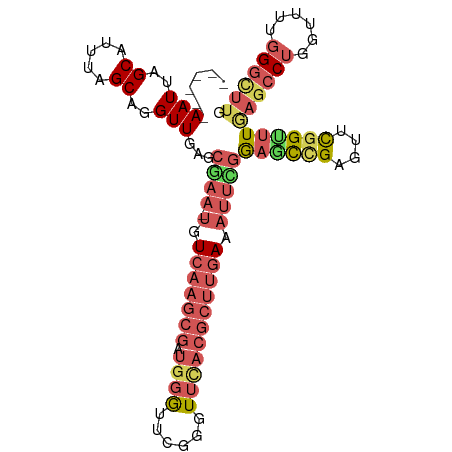
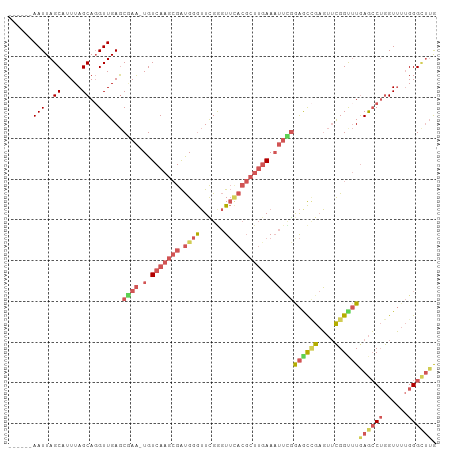
| Location | 3,268,130 – 3,268,222 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | forward |
| Mean pairwise identity | 94.93 |
| Shannon entropy | 0.08142 |
| G+C content | 0.46196 |
| Mean single sequence MFE | -22.06 |
| Consensus MFE | -20.70 |
| Energy contribution | -20.76 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.94 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.951384 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
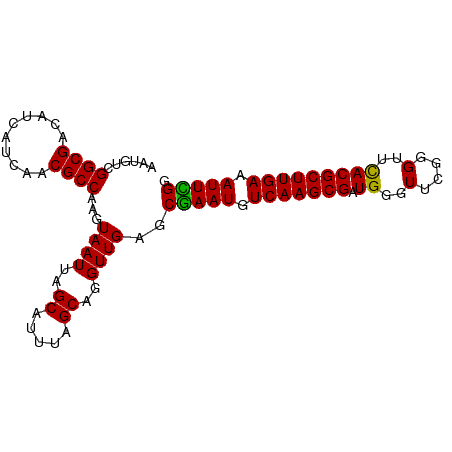
>dm3.chr3R 3268130 92 + 27905053 CCAAAUUUCAAGCGUAAACCCAAACCCAUCGCUUGACAUUUGCUCAACAUGCUAAAUGCUAAUUACUUGGCGUUGAUGAUGUCGCCGACAUU .(((((.(((((((...............))))))).))))).(((((((.....))(((((....)))))))))).((((((...)))))) ( -21.36, z-score = -2.47, R) >droSim1.chr3R 3312555 92 + 27517382 CCGAAUUUCAAGCGUGAACCCGAACCCAUCGCUUGACAUUCGCUCAACCUGCUAAAUGCUAAUUACUUGGCGUUGAUGAUGUCGCCGACAUU .(((((.(((((((((..........)).))))))).)))))............((((((((....))))))))...((((((...)))))) ( -24.00, z-score = -2.77, R) >droSec1.super_6 3359246 92 + 4358794 CCGAAUUUCAAGCGUGAACCCGAACCCAUCGCUUGACAUUCGCUCAACCUGCUAAAUGCUAAUUACUUGGCGUUGAUGAUGUCGCCGACAUU .(((((.(((((((((..........)).))))))).)))))............((((((((....))))))))...((((((...)))))) ( -24.00, z-score = -2.77, R) >droYak2.chr3R_random 962847 92 + 3277457 UCAAAUUUCAAGCGUGAACUCGAACCCAUCGCUUGACAUUCGCUCAACCUGCUAAAUGCUAAUUACUUGGCGUCGAUGGUGUCGCCGACAUU .......(((((((((..........)).)))))))((((.((.......))..))))........((((((.((....)).)))))).... ( -18.90, z-score = -0.25, R) >consensus CCAAAUUUCAAGCGUGAACCCGAACCCAUCGCUUGACAUUCGCUCAACCUGCUAAAUGCUAAUUACUUGGCGUUGAUGAUGUCGCCGACAUU .(((((.(((((((...............))))))).)))))............((((((((....))))))))...((((((...)))))) (-20.70 = -20.76 + 0.06)
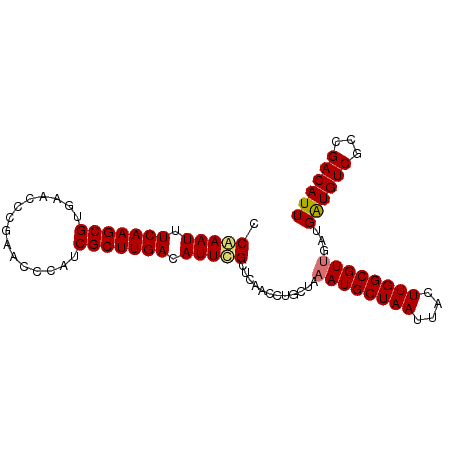
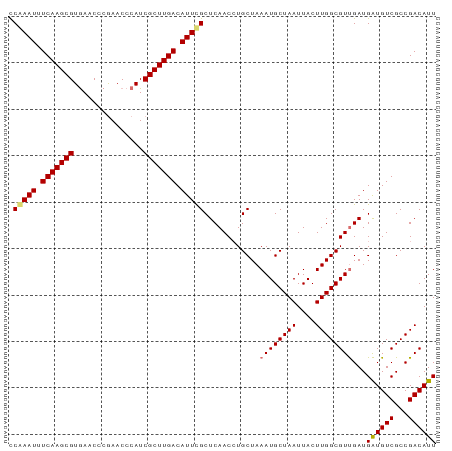
| Location | 3,268,130 – 3,268,222 |
|---|---|
| Length | 92 |
| Sequences | 4 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 94.93 |
| Shannon entropy | 0.08142 |
| G+C content | 0.46196 |
| Mean single sequence MFE | -25.68 |
| Consensus MFE | -25.42 |
| Energy contribution | -24.80 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.99 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.838444 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 3268130 92 - 27905053 AAUGUCGGCGACAUCAUCAACGCCAAGUAAUUAGCAUUUAGCAUGUUGAGCAAAUGUCAAGCGAUGGGUUUGGGUUUACGCUUGAAAUUUGG ......((((..........))))......(((((((.....))))))).(((((.(((((((.((..(....)..))))))))).))))). ( -23.00, z-score = -0.72, R) >droSim1.chr3R 3312555 92 - 27517382 AAUGUCGGCGACAUCAUCAACGCCAAGUAAUUAGCAUUUAGCAGGUUGAGCGAAUGUCAAGCGAUGGGUUCGGGUUCACGCUUGAAAUUCGG ......((((..........))))...((((..((.....))..))))..(((((.(((((((.((..(....)..))))))))).))))). ( -27.10, z-score = -1.74, R) >droSec1.super_6 3359246 92 - 4358794 AAUGUCGGCGACAUCAUCAACGCCAAGUAAUUAGCAUUUAGCAGGUUGAGCGAAUGUCAAGCGAUGGGUUCGGGUUCACGCUUGAAAUUCGG ......((((..........))))...((((..((.....))..))))..(((((.(((((((.((..(....)..))))))))).))))). ( -27.10, z-score = -1.74, R) >droYak2.chr3R_random 962847 92 - 3277457 AAUGUCGGCGACACCAUCGACGCCAAGUAAUUAGCAUUUAGCAGGUUGAGCGAAUGUCAAGCGAUGGGUUCGAGUUCACGCUUGAAAUUUGA ......((((.(......).))))...((((..((.....))..))))..(((((.(((((((.((..(....)..))))))))).))))). ( -25.50, z-score = -0.74, R) >consensus AAUGUCGGCGACAUCAUCAACGCCAAGUAAUUAGCAUUUAGCAGGUUGAGCGAAUGUCAAGCGAUGGGUUCGGGUUCACGCUUGAAAUUCGG ......((((..........))))...((((..((.....))..))))..(((((.(((((((.((..(....)..))))))))).))))). (-25.42 = -24.80 + -0.62)

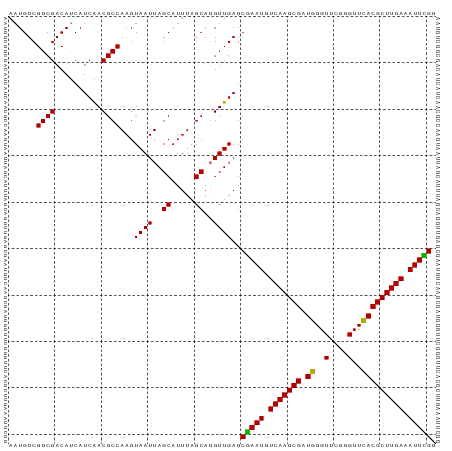
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:56:13 2011