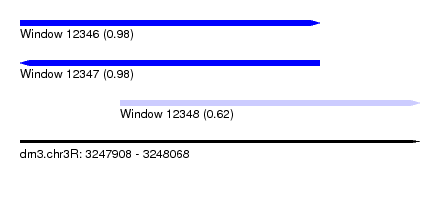
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 3,247,908 – 3,248,068 |
| Length | 160 |
| Max. P | 0.980253 |

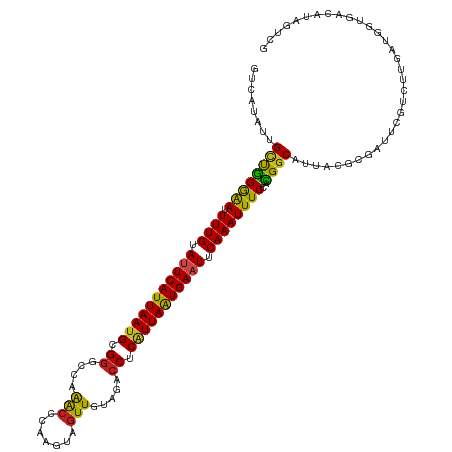
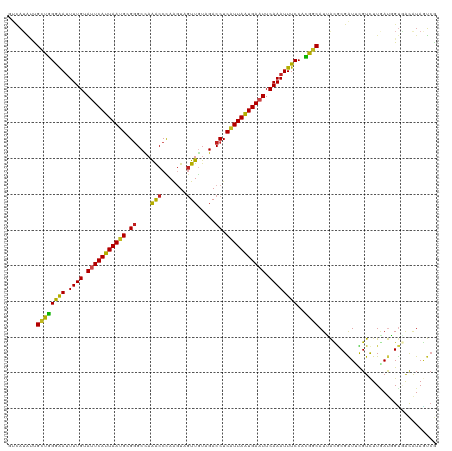
| Location | 3,247,908 – 3,248,028 |
|---|---|
| Length | 120 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.20 |
| Shannon entropy | 0.32790 |
| G+C content | 0.39594 |
| Mean single sequence MFE | -31.52 |
| Consensus MFE | -17.62 |
| Energy contribution | -16.92 |
| Covariance contribution | -0.71 |
| Combinations/Pair | 1.38 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.04 |
| SVM RNA-class probability | 0.980253 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
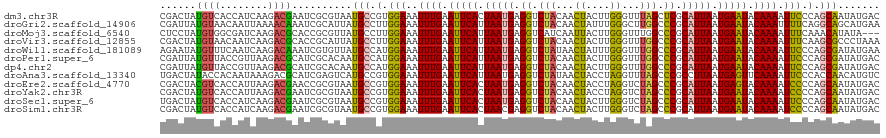
>dm3.chr3R 3247908 120 + 27905053 CGACUAUGUCACCAUCAAGACGAAUCGCGUAAUGCCGUGGAAAUUUGAAUUCACUAAUGAGGUCUACAACUACUUGGGUUUAGCUCGCAUUAAUGAAUACAAAAUUCCCAGCAAUAUGAC (((...((((........))))..)))((((.(((.(.((((.((((.(((((.(((((.(..(((.((((.....)))))))..).))))).))))).)))).))))).))).)))).. ( -30.90, z-score = -2.78, R) >droGri2.scaffold_14906 6222942 120 - 14172833 CGAUUAUGUAACAAUUAAAACAAAUCGCAUUAUGCCUUGGAAAUUUGAAUUCAUUAAUGAGGUCUACAACUAUUUGGGCUUGGCCCGCAUUAAUGAAUACAAAUUUUCAGGCAGCAUGAA (((((.(((..........))))))))..((((((((((((((((((.(((((((((((((........))....((((...)))).))))))))))).))))))))))))..)))))). ( -38.70, z-score = -5.37, R) >droMoj3.scaffold_6540 6459907 117 + 34148556 CUCCUAUGUGGCGAUCAAGACGCACCGCGUUAUGCCUUGGAAAUUUGAAUUCAUUAAUGAGGUCAUCAAUUACUUGGGUUUGGCCCGCAUUAAUGAAUACAAAAUUUCAAACAUAUA--- ....(((((((((.....(((((...))))).))))((((((.((((.(((((((((((.(((((((((....))))...)))))..))))))))))).)))).)))))))))))..--- ( -35.20, z-score = -3.48, R) >droVir3.scaffold_12855 6130390 120 + 10161210 CGACUAUGUAACAAUCAAGACGCACCGCAUUAUGCCUUGGAAAUUUGAAUUCAUUAAUGAGGUCUACAACUACUUGGGUUUGGCCCGCAUUAAUGAAUACAAAAUUUCAAGCGCCCUAAA .................................(((((((((.((((.(((((((((((.((.(((.((((.....))))))).)).))))))))))).)))).))))))).))...... ( -30.90, z-score = -2.88, R) >droWil1.scaffold_181089 7128130 120 + 12369635 AGAAUAUGUUUCAAUCAAGACAAAUCGUGUUAUGCCAUGGAAAUUUGAAUUCAUUAAUGAGGUCUAUAACUAUUUGGGUUUGGCCCGCAUUAAUGAAUACAAAAUUCCCAGCGAUAUGAA ......(((((......)))))..(((((((..((...((((.((((.(((((((((((.((((...((((.....)))).))))..))))))))))).)))).))))..))))))))). ( -34.40, z-score = -3.53, R) >droPer1.super_6 1968783 120 + 6141320 CGAUUAUGUUACCGUUAAGACGCAUCGCACAAUGCCAUGGAAAUUUGAAUUCAUUAAUGAGGUCUACAACUACUUGGGUUUGGCCCGCAUUAAUGAAUACAAAAUUCCCAGCGAUAUGAC ...((((((...((((.....((((......))))...((((.((((.(((((((((((.((.(((.((((.....))))))).)).))))))))))).)))).)))).)))))))))). ( -32.10, z-score = -2.67, R) >dp4.chr2 26640148 120 + 30794189 CGAUUAUGUUACCGUUAAGACGCAUCGCACAAUGCCAUGGAAAUUUGAAUUCAUUAAUGAGGUCUACAACUACUUGGGUUUGGCCCGCAUUAAUGAAUACAAAAUUCCCAGCGAUAUGAC ...((((((...((((.....((((......))))...((((.((((.(((((((((((.((.(((.((((.....))))))).)).))))))))))).)))).)))).)))))))))). ( -32.10, z-score = -2.67, R) >droAna3.scaffold_13340 3033498 120 + 23697760 UGACUAUACCACAAUAAAGACGCAUCGAGUCAUGCCGUGGAAAUUUGAAUUCAUUAAUGAGGUCUAUAACUACCUAGGUUUAGCCCGCCUUAAUGAGUUCAAAAUUCCCACCAACAUGUC ..................(((((((......)))).((((((.((((((((((((((.(.((.(((.((((.....))))))).)).).)))))))))))))).)).))))......))) ( -31.20, z-score = -3.58, R) >droEre2.scaffold_4770 3512274 120 + 17746568 CGACUACGUCACCAUUAAGACGAACCGCGUAAUGCCGUGGAAAUUUGAAUUCACUAAUGAGGUCUACAACUACCUAGGUCUAGCCCGCAUUAAUGAGUACAAAAUCCCCAGCAAUAUGAC ......((((........)))).....((((.(((.(.(((..((((.(((((.(((((.((.(((..(((.....))).))).)).))))).))))).)))).))).).))).)))).. ( -29.50, z-score = -2.72, R) >droYak2.chr3R 21731200 120 - 28832112 CGACUAUGUCACCAUUAAGACGAAUCGCGUAAUGCCGUGGAAAUUUGAAUUCACUAAUGAGGUCUACAACUACCUAGGUCUAGCCCGCAUUAAUGAAUACAAAAUCCCCAGCAAUAUGAC (((...((((........))))..)))((((.(((.(.(((..((((.(((((.(((((.((.(((..(((.....))).))).)).))))).))))).)))).))).).))).)))).. ( -28.40, z-score = -2.65, R) >droSec1.super_6 3339237 120 + 4358794 UGACUAUGUCACCAUCAAGACGAAUCGCGUAAUGCCGUGGAAAUUUGAAUUCACUAAUGAGGUCUACAACUACUUGGGUCUAGCCCGCAUUAAUGAAUACAAAAUUCCCAGCAAUAUGAC ......((((........)))).....((((.(((.(.((((.((((.(((((.(((((.((.(((..(((.....))).))).)).))))).))))).)))).))))).))).)))).. ( -30.70, z-score = -2.64, R) >droSim1.chr3R 3292320 120 + 27517382 CGACUAUGUCACCAUCAAGACGAAUCGCGUAAUGCCGUGGAAAUUUGAAUUCACUAACGAGGUCUACAACUACUUGGGUCUAGCCCGCAUUAAUGAAUACAAAAUCCCCAGCAAUAUGAC (((...((((........))))..)))((((.(((.(.(((..((((.(((((.(((.(.((.(((..(((.....))).))).)).).))).))))).)))).))).).))).)))).. ( -24.20, z-score = -0.63, R) >consensus CGACUAUGUCACCAUCAAGACGAAUCGCGUAAUGCCGUGGAAAUUUGAAUUCAUUAAUGAGGUCUACAACUACUUGGGUUUAGCCCGCAUUAAUGAAUACAAAAUUCCCAGCAAUAUGAC ......((((........))))..........(((.(.(((..((((.(((((.(((((.((.(((...((....))...))).)).))))).))))).)))).))).).)))....... (-17.62 = -16.92 + -0.71)

| Location | 3,247,908 – 3,248,028 |
|---|---|
| Length | 120 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 85.20 |
| Shannon entropy | 0.32790 |
| G+C content | 0.39594 |
| Mean single sequence MFE | -34.07 |
| Consensus MFE | -22.24 |
| Energy contribution | -21.11 |
| Covariance contribution | -1.13 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.99 |
| SVM RNA-class probability | 0.978046 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 3247908 120 - 27905053 GUCAUAUUGCUGGGAAUUUUGUAUUCAUUAAUGCGAGCUAAACCCAAGUAGUUGUAGACCUCAUUAGUGAAUUCAAAUUUCCACGGCAUUACGCGAUUCGUCUUGAUGGUGACAUAGUCG (((((..(((((((((.((((.(((((((((((.(..((((((.......))).)))..).))))))))))).)))).)))).)))))..........(((....))))))))....... ( -35.40, z-score = -3.09, R) >droGri2.scaffold_14906 6222942 120 + 14172833 UUCAUGCUGCCUGAAAAUUUGUAUUCAUUAAUGCGGGCCAAGCCCAAAUAGUUGUAGACCUCAUUAAUGAAUUCAAAUUUCCAAGGCAUAAUGCGAUUUGUUUUAAUUGUUACAUAAUCG ..(((..(((((..(((((((.(((((((((((.((((...)))).....(((...)))..))))))))))).)))))))...)))))..)))((((((((..........))).))))) ( -32.40, z-score = -3.59, R) >droMoj3.scaffold_6540 6459907 117 - 34148556 ---UAUAUGUUUGAAAUUUUGUAUUCAUUAAUGCGGGCCAAACCCAAGUAAUUGAUGACCUCAUUAAUGAAUUCAAAUUUCCAAGGCAUAACGCGGUGCGUCUUGAUCGCCACAUAGGAG ---..((((((((((((((.(.(((((((((((.(((.((....(((....))).)).)))))))))))))).))))))))..)))))))..(((((........))))).......... ( -29.30, z-score = -1.72, R) >droVir3.scaffold_12855 6130390 120 - 10161210 UUUAGGGCGCUUGAAAUUUUGUAUUCAUUAAUGCGGGCCAAACCCAAGUAGUUGUAGACCUCAUUAAUGAAUUCAAAUUUCCAAGGCAUAAUGCGGUGCGUCUUGAUUGUUACAUAGUCG ...(((((.((((.((.((((.(((((((((((.((....(((.......))).....)).))))))))))).)))).)).))))((((......)))))))))((((((...)))))). ( -32.60, z-score = -2.12, R) >droWil1.scaffold_181089 7128130 120 - 12369635 UUCAUAUCGCUGGGAAUUUUGUAUUCAUUAAUGCGGGCCAAACCCAAAUAGUUAUAGACCUCAUUAAUGAAUUCAAAUUUCCAUGGCAUAACACGAUUUGUCUUGAUUGAAACAUAUUCU ((((.(((((((((((.((((.(((((((((((.((....(((.......))).....)).))))))))))).)))).)))).)))).......((....))..)))))))......... ( -28.90, z-score = -2.68, R) >droPer1.super_6 1968783 120 - 6141320 GUCAUAUCGCUGGGAAUUUUGUAUUCAUUAAUGCGGGCCAAACCCAAGUAGUUGUAGACCUCAUUAAUGAAUUCAAAUUUCCAUGGCAUUGUGCGAUGCGUCUUAACGGUAACAUAAUCG ........((((((((.((((.(((((((((((.((....(((.......))).....)).))))))))))).)))).))))...(((((....))))).......)))).......... ( -32.80, z-score = -2.30, R) >dp4.chr2 26640148 120 - 30794189 GUCAUAUCGCUGGGAAUUUUGUAUUCAUUAAUGCGGGCCAAACCCAAGUAGUUGUAGACCUCAUUAAUGAAUUCAAAUUUCCAUGGCAUUGUGCGAUGCGUCUUAACGGUAACAUAAUCG ........((((((((.((((.(((((((((((.((....(((.......))).....)).))))))))))).)))).))))...(((((....))))).......)))).......... ( -32.80, z-score = -2.30, R) >droAna3.scaffold_13340 3033498 120 - 23697760 GACAUGUUGGUGGGAAUUUUGAACUCAUUAAGGCGGGCUAAACCUAGGUAGUUAUAGACCUCAUUAAUGAAUUCAAAUUUCCACGGCAUGACUCGAUGCGUCUUUAUUGUGGUAUAGUCA (((((((((...((((.((((((.(((((((...(((((((((.......))).))).)))..))))))).)))))).)))).)))))).((((((((......))))).)))...))). ( -34.10, z-score = -2.12, R) >droEre2.scaffold_4770 3512274 120 - 17746568 GUCAUAUUGCUGGGGAUUUUGUACUCAUUAAUGCGGGCUAGACCUAGGUAGUUGUAGACCUCAUUAGUGAAUUCAAAUUUCCACGGCAUUACGCGGUUCGUCUUAAUGGUGACGUAGUCG ....((.(((((((((.((((...(((((((((.((.(((.(.((....)).).))).)).)))))))))...)))).)))).))))).))..((..(((((........)))).)..)) ( -36.90, z-score = -2.33, R) >droYak2.chr3R 21731200 120 + 28832112 GUCAUAUUGCUGGGGAUUUUGUAUUCAUUAAUGCGGGCUAGACCUAGGUAGUUGUAGACCUCAUUAGUGAAUUCAAAUUUCCACGGCAUUACGCGAUUCGUCUUAAUGGUGACAUAGUCG (((((..(((((((((.((((.(((((((((((.((.(((.(.((....)).).))).)).))))))))))).)))).)))).)))))..(((.....))).......)))))....... ( -38.10, z-score = -3.31, R) >droSec1.super_6 3339237 120 - 4358794 GUCAUAUUGCUGGGAAUUUUGUAUUCAUUAAUGCGGGCUAGACCCAAGUAGUUGUAGACCUCAUUAGUGAAUUCAAAUUUCCACGGCAUUACGCGAUUCGUCUUGAUGGUGACAUAGUCA (((((..(((((((((.((((.(((((((((((.((.((((((.......))).))).)).))))))))))).)))).)))).)))))..........(((....))))))))....... ( -38.20, z-score = -3.30, R) >droSim1.chr3R 3292320 120 - 27517382 GUCAUAUUGCUGGGGAUUUUGUAUUCAUUAAUGCGGGCUAGACCCAAGUAGUUGUAGACCUCGUUAGUGAAUUCAAAUUUCCACGGCAUUACGCGAUUCGUCUUGAUGGUGACAUAGUCG (((((..(((((((((.((((.(((((((((((.((.((((((.......))).))).)).))))))))))).)))).)))).)))))..........(((....))))))))....... ( -37.30, z-score = -2.56, R) >consensus GUCAUAUUGCUGGGAAUUUUGUAUUCAUUAAUGCGGGCCAAACCCAAGUAGUUGUAGACCUCAUUAAUGAAUUCAAAUUUCCACGGCAUUACGCGAUUCGUCUUGAUGGUGACAUAGUCG ........((((((((.((((.(((((((((((.((....(((.......))).....)).))))))))))).))))))))..))))................................. (-22.24 = -21.11 + -1.13)
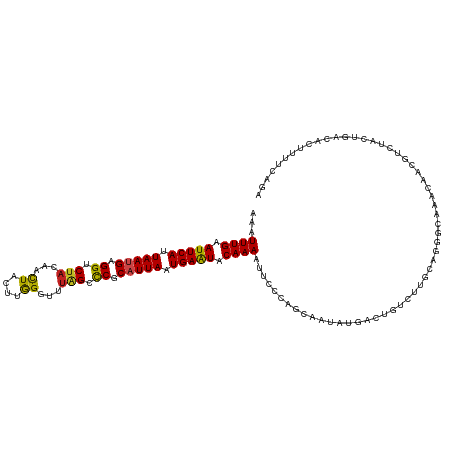
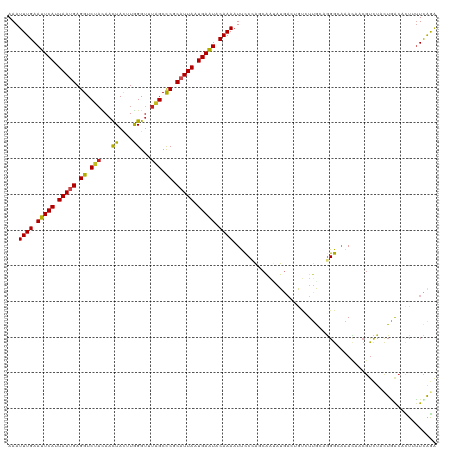
| Location | 3,247,948 – 3,248,068 |
|---|---|
| Length | 120 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.44 |
| Shannon entropy | 0.41006 |
| G+C content | 0.37990 |
| Mean single sequence MFE | -26.49 |
| Consensus MFE | -14.49 |
| Energy contribution | -14.14 |
| Covariance contribution | -0.35 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.26 |
| SVM RNA-class probability | 0.618228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
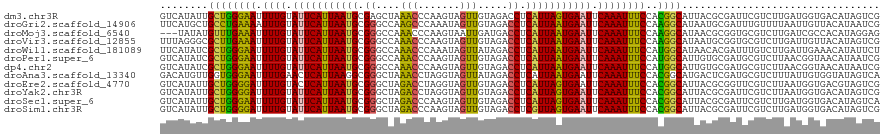
>dm3.chr3R 3247948 120 + 27905053 AAAUUUGAAUUCACUAAUGAGGUCUACAACUACUUGGGUUUAGCUCGCAUUAAUGAAUACAAAAUUCCCAGCAAUAUGACUGUCUUGCAGGGCAAACAACGUCUGCUAACACUUUUUAGA ...((((.(((((.(((((.(..(((.((((.....)))))))..).))))).))))).))))...(((.((((.((....)).)))).))).............((((......)))). ( -22.50, z-score = -0.51, R) >droGri2.scaffold_14906 6222982 120 - 14172833 AAAUUUGAAUUCAUUAAUGAGGUCUACAACUAUUUGGGCUUGGCCCGCAUUAAUGAAUACAAAUUUUCAGGCAGCAUGAAAACGUUAACUGGGAGAAAACGCUUACUAACAUUGUUCAAU ...((((.(((((((((((((........))....((((...)))).))))))))))).))))((..(((..(((.(....).)))..)))..))......................... ( -27.40, z-score = -2.16, R) >droMoj3.scaffold_6540 6459947 117 + 34148556 AAAUUUGAAUUCAUUAAUGAGGUCAUCAAUUACUUGGGUUUGGCCCGCAUUAAUGAAUACAAAAUUUCAAACA---UAUACAAAUCGAUGGGGCGAUCGAAUCUGCUGGAUCUAUUUAGG ...((((.(((((((((((.(((((((((....))))...)))))..))))))))))).))))..........---.....((((.(((..((((........))))..))).))))... ( -26.20, z-score = -1.53, R) >droVir3.scaffold_12855 6130430 120 + 10161210 AAAUUUGAAUUCAUUAAUGAGGUCUACAACUACUUGGGUUUGGCCCGCAUUAAUGAAUACAAAAUUUCAAGCGCCCUAAAAACAUUAUCGGGACGAACACGUCUACUGAAUUUACUCAAG ...((((.(((((((((((.((.(((.((((.....))))))).)).))))))))))).))))........................((((((((....))))..))))........... ( -26.90, z-score = -2.74, R) >droWil1.scaffold_181089 7128170 120 + 12369635 AAAUUUGAAUUCAUUAAUGAGGUCUAUAACUAUUUGGGUUUGGCCCGCAUUAAUGAAUACAAAAUUCCCAGCGAUAUGAAAGUCUUGCAGGGUAAACAACGUCUCCUAACACUUUUCAAA ...((((.(((((((((((.((((...((((.....)))).))))..))))))))))).))))...(((.((((.((....)).)))).)))............................ ( -30.40, z-score = -2.89, R) >droPer1.super_6 1968823 120 + 6141320 AAAUUUGAAUUCAUUAAUGAGGUCUACAACUACUUGGGUUUGGCCCGCAUUAAUGAAUACAAAAUUCCCAGCGAUAUGACCGUCUUAAAGGGCAAACAACGUCUAUUGACACUCUUCGGA ...((((.(((((((((((.((.(((.((((.....))))))).)).))))))))))).))))................((((((....)))).......(((....))).......)). ( -29.10, z-score = -1.91, R) >dp4.chr2 26640188 120 + 30794189 AAAUUUGAAUUCAUUAAUGAGGUCUACAACUACUUGGGUUUGGCCCGCAUUAAUGAAUACAAAAUUCCCAGCGAUAUGACCGUCUUAAAGGGCAAACAACGUCUAUUGACACUCUUUGGA ...((((.(((((((((((.((.(((.((((.....))))))).)).))))))))))).))))....((((.((.......((((....)))).......(((....)))..)).)))). ( -31.10, z-score = -2.59, R) >droAna3.scaffold_13340 3033538 120 + 23697760 AAAUUUGAAUUCAUUAAUGAGGUCUAUAACUACCUAGGUUUAGCCCGCCUUAAUGAGUUCAAAAUUCCCACCAACAUGUCGAUCUUGCAAGGCAAACAACGUCUCCUAACCUUAUUUAGA ...((((((((((((((.(.((.(((.((((.....))))))).)).).)))))))))))))).............((((..........))))...........((((......)))). ( -23.90, z-score = -2.10, R) >droEre2.scaffold_4770 3512314 120 + 17746568 AAAUUUGAAUUCACUAAUGAGGUCUACAACUACCUAGGUCUAGCCCGCAUUAAUGAGUACAAAAUCCCCAGCAAUAUGACUGUCUUGCAGGGAAGACAACGUCUUCUGACACUUUUCAGA ...((((.(((((.(((((.((.(((..(((.....))).))).)).))))).))))).)))).((((..((((.((....)).)))).))))((((...))))(((((......))))) ( -28.60, z-score = -1.60, R) >droYak2.chr3R 21731240 120 - 28832112 AAAUUUGAAUUCACUAAUGAGGUCUACAACUACCUAGGUCUAGCCCGCAUUAAUGAAUACAAAAUCCCCAGCAAUAUGACUGUCUUGCAGGGAAGACAACGUCUUCUGACACUUUUCAGA ...((((.(((((.(((((.((.(((..(((.....))).))).)).))))).))))).)))).((((..((((.((....)).)))).))))((((...))))(((((......))))) ( -28.60, z-score = -2.08, R) >droSec1.super_6 3339277 120 + 4358794 AAAUUUGAAUUCACUAAUGAGGUCUACAACUACUUGGGUCUAGCCCGCAUUAAUGAAUACAAAAUUCCCAGCAAUAUGACUGUCUUGCAGGGCAAACAACGUCUGCUAACCCUUUUCAGA ...((((.(((((.(((((.((.(((..(((.....))).))).)).))))).))))).))))......((((....((((((.((((...)))))))..)))))))............. ( -23.00, z-score = -0.22, R) >droSim1.chr3R 3292360 120 + 27517382 AAAUUUGAAUUCACUAACGAGGUCUACAACUACUUGGGUCUAGCCCGCAUUAAUGAAUACAAAAUCCCCAGCAAUAUGACUGUCUUGCAGGGCAAACAACGUCUGCUCACACUUUUCAGA ...((((.(((((.(((.(.((.(((..(((.....))).))).)).).))).))))).)))).......((((.((....)).)))).(((((.((...)).)))))............ ( -20.20, z-score = 0.59, R) >consensus AAAUUUGAAUUCAUUAAUGAGGUCUACAACUACUUGGGUUUAGCCCGCAUUAAUGAAUACAAAAUUCCCAGCAAUAUGACUGUCUUGCAGGGCAAACAACGUCUACUGACACUUUUCAGA ...((((.(((((.(((((.((.(((...((....))...))).)).))))).))))).))))......................................................... (-14.49 = -14.14 + -0.35)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:56:08 2011