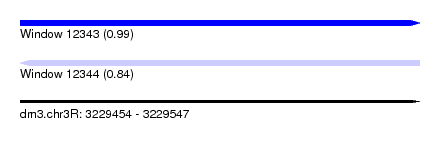
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 3,229,454 – 3,229,547 |
| Length | 93 |
| Max. P | 0.994196 |

| Location | 3,229,454 – 3,229,547 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 74.03 |
| Shannon entropy | 0.52313 |
| G+C content | 0.43060 |
| Mean single sequence MFE | -22.51 |
| Consensus MFE | -10.78 |
| Energy contribution | -11.39 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.65 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.68 |
| SVM RNA-class probability | 0.994196 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
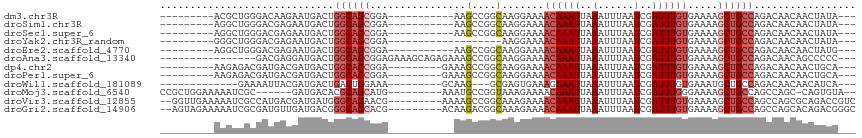
>dm3.chr3R 3229454 93 + 27905053 ---UAUAGUUGUUGUCUGGCAGCUUUUCACAAAUCGAUUAAAUUUAAUUUGUUUUCCUUGCCGGCUU-----------UCCGCUGCCAGUCAUUCUUGUCCCAGCGU--------- ---....((((.((.((((((((.....((((((..(......)..))))))..........((...-----------.)))))))))).)).........))))..--------- ( -23.30, z-score = -3.16, R) >droSim1.chr3R 3273666 93 + 27517382 ---UAUAGUUGUUGUCUGGCAGCUUUUCACAAAUCGAUUAAAUUUAAUUUGUUUUCCUUGCCGGCUU-----------UCCGCUGCCAGUCAUUCUCGUCCCAGCCU--------- ---....((((.((.((((((((.....((((((..(......)..))))))..........((...-----------.)))))))))).)).........))))..--------- ( -22.40, z-score = -3.22, R) >droSec1.super_6 3320976 93 + 4358794 ---UAUAGUUGUUGUCUGGCAGCUUUUCACAAAUCGAUUAAAUUUAAUUUGUUUUCCUUGCCGGCUU-----------UCCGCUGCCAGUCAUUCUCGUCCCAGCCU--------- ---....((((.((.((((((((.....((((((..(......)..))))))..........((...-----------.)))))))))).)).........))))..--------- ( -22.40, z-score = -3.22, R) >droYak2.chr3R_random 928577 85 + 3277457 ---UAUAGUUGUUGUCUGGCAGCUUUUCACAAAUCGAUUAAAUUUAAUUUGUUUUCCUU-------------------UCCGCUGCCAGUCAUUCUCGUCCCAGCCC--------- ---....((((.((.((((((((.....((((((..(......)..)))))).......-------------------...)))))))).)).........))))..--------- ( -19.26, z-score = -4.26, R) >droEre2.scaffold_4770 3488387 93 + 17746568 ---CAUAGUUGUUGUCUGGCAGCUUUUCACAAAUCGAUUAAAUUUAAUUUGUUUUCCUUGCCGGCUU-----------UCCGCUGCCAGUCAUUCUCGUCCCAGCCU--------- ---....((((.((.((((((((.....((((((..(......)..))))))..........((...-----------.)))))))))).)).........))))..--------- ( -22.40, z-score = -3.15, R) >droAna3.scaffold_13340 3016637 97 + 23697760 ---GGGGGCUGUUGUCUGGCAGCUUUUCACAAAUCGAUUAAAUUUAAUUUGUUUUCCUUGCCGGCUUUCUCUGCUUUCUCCGCUGCCAGUCAUCCUCGUC---------------- ---(((((....((.((((((((.....((((((..(......)..))))))..........(((.......)))......)))))))).)))))))...---------------- ( -28.50, z-score = -3.52, R) >dp4.chr2 26622753 95 + 30794189 ---UGCAGUUGUUGUCUGGCAGCUUUUCACAAAUCGAUUAAAUUUAAUUUGUUUUCCUUGCCGGCUUUC---------UCCGCUGCCAGUCAUCGUCAUCGUCUCUU--------- ---....(.((.((.((((((((.....((((((..(......)..))))))..........((.....---------.)))))))))).)).)).)..........--------- ( -23.10, z-score = -3.25, R) >droPer1.super_6 1951328 95 + 6141320 ---UGCAGUUGUUGUCUGGCAGCUUUUCACAAAUCGAUUAAAUUUAAUUUGUUUUCCUUGCCGGCUUUC---------UCCGCUGCCAGUCAUCGUCAUCGUCUCUU--------- ---....(.((.((.((((((((.....((((((..(......)..))))))..........((.....---------.)))))))))).)).)).)..........--------- ( -23.10, z-score = -3.25, R) >droWil1.scaffold_181089 7110223 88 + 12369635 ---UGAUGUUGUUGUCUGGCAGCAUUUCACAAAUCGAUUAAAUUUAAUUUCUUUCACUCGC---CUUGC---------UUUCCAGUCAGUCAUCGUAAUUUUC------------- ---.((((...(((.((((.((((...........(((((....)))))............---..)))---------)..)))).))).)))).........------------- ( -13.71, z-score = -1.05, R) >droMoj3.scaffold_6540 6436029 98 + 34148556 --UACACUG-GCUGGCUGGCAGCUUUUCCCAAAUCGAUUAAAUUUAAUUUGUUUUCUUUACCGGCAUUU---------CAUGCUGCGUGUCAUC------GCGAUUUUUCCAGCGG --.......-(((((...(((((......(((((..(......)..)))))...........((....)---------)..)))))(((....)------)).......))))).. ( -19.80, z-score = 0.25, R) >droVir3.scaffold_12855 6107925 105 + 10161210 GACGGUCUGCGCUGGCUGGCAGCUUUUCACAAAUCGAUUAAAUUUAAUUUGUUUUCUUUGCCGGCUUUU---------CGUUCUGCCCAUCAUCGUCAUGGCGAUUUUUCAACC-- ...(((..(((..(((((((((......((((((..(......)..)))))).....)))))))))...---------)))...)))....(((((....))))).........-- ( -28.20, z-score = -2.19, R) >droGri2.scaffold_14906 6211032 105 - 14172833 GCCCGUCUGUGCUGGCUGGCAGCUUUUCACAAAUCGAUUAAAUUUAAUUUGUUUUCUUUGCCGUCUUGU---------CGUGCUGCCCGUCAUCAACAUCGCGAUUUUUCUACU-- ...(((.(((..((((.((((((.....((((((..(......)..)))))).........((......---------)).)))))).))))...)))..)))...........-- ( -23.90, z-score = -1.76, R) >consensus ___UGUAGUUGUUGUCUGGCAGCUUUUCACAAAUCGAUUAAAUUUAAUUUGUUUUCCUUGCCGGCUU___________UCCGCUGCCAGUCAUCCUCGUCCCAGCUU_________ ............((.(.((((((.....((((((..(......)..)))))).......(....)................)))))).).))........................ (-10.78 = -11.39 + 0.61)

| Location | 3,229,454 – 3,229,547 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 74.03 |
| Shannon entropy | 0.52313 |
| G+C content | 0.43060 |
| Mean single sequence MFE | -21.16 |
| Consensus MFE | -10.72 |
| Energy contribution | -11.25 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.836748 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 3229454 93 - 27905053 ---------ACGCUGGGACAAGAAUGACUGGCAGCGGA-----------AAGCCGGCAAGGAAAACAAAUUAAAUUUAAUCGAUUUGUGAAAAGCUGCCAGACAACAACUAUA--- ---------...............((.((((((((...-----------...((.....))...((((((..(......)..)))))).....)))))))).)).........--- ( -22.00, z-score = -2.17, R) >droSim1.chr3R 3273666 93 - 27517382 ---------AGGCUGGGACGAGAAUGACUGGCAGCGGA-----------AAGCCGGCAAGGAAAACAAAUUAAAUUUAAUCGAUUUGUGAAAAGCUGCCAGACAACAACUAUA--- ---------...............((.((((((((...-----------...((.....))...((((((..(......)..)))))).....)))))))).)).........--- ( -22.00, z-score = -1.89, R) >droSec1.super_6 3320976 93 - 4358794 ---------AGGCUGGGACGAGAAUGACUGGCAGCGGA-----------AAGCCGGCAAGGAAAACAAAUUAAAUUUAAUCGAUUUGUGAAAAGCUGCCAGACAACAACUAUA--- ---------...............((.((((((((...-----------...((.....))...((((((..(......)..)))))).....)))))))).)).........--- ( -22.00, z-score = -1.89, R) >droYak2.chr3R_random 928577 85 - 3277457 ---------GGGCUGGGACGAGAAUGACUGGCAGCGGA-------------------AAGGAAAACAAAUUAAAUUUAAUCGAUUUGUGAAAAGCUGCCAGACAACAACUAUA--- ---------...............((.((((((((...-------------------.......((((((..(......)..)))))).....)))))))).)).........--- ( -19.06, z-score = -2.48, R) >droEre2.scaffold_4770 3488387 93 - 17746568 ---------AGGCUGGGACGAGAAUGACUGGCAGCGGA-----------AAGCCGGCAAGGAAAACAAAUUAAAUUUAAUCGAUUUGUGAAAAGCUGCCAGACAACAACUAUG--- ---------...............((.((((((((...-----------...((.....))...((((((..(......)..)))))).....)))))))).)).........--- ( -22.00, z-score = -1.84, R) >droAna3.scaffold_13340 3016637 97 - 23697760 ----------------GACGAGGAUGACUGGCAGCGGAGAAAGCAGAGAAAGCCGGCAAGGAAAACAAAUUAAAUUUAAUCGAUUUGUGAAAAGCUGCCAGACAACAGCCCCC--- ----------------.....((.((.((((((((.................((.....))...((((((..(......)..)))))).....)))))))).))....))...--- ( -22.80, z-score = -1.81, R) >dp4.chr2 26622753 95 - 30794189 ---------AAGAGACGAUGACGAUGACUGGCAGCGGA---------GAAAGCCGGCAAGGAAAACAAAUUAAAUUUAAUCGAUUUGUGAAAAGCUGCCAGACAACAACUGCA--- ---------.............(.((.((((((((((.---------.....))..........((((((..(......)..)))))).....)))))))).)).).......--- ( -22.30, z-score = -2.33, R) >droPer1.super_6 1951328 95 - 6141320 ---------AAGAGACGAUGACGAUGACUGGCAGCGGA---------GAAAGCCGGCAAGGAAAACAAAUUAAAUUUAAUCGAUUUGUGAAAAGCUGCCAGACAACAACUGCA--- ---------.............(.((.((((((((((.---------.....))..........((((((..(......)..)))))).....)))))))).)).).......--- ( -22.30, z-score = -2.33, R) >droWil1.scaffold_181089 7110223 88 - 12369635 -------------GAAAAUUACGAUGACUGACUGGAAA---------GCAAG---GCGAGUGAAAGAAAUUAAAUUUAAUCGAUUUGUGAAAUGCUGCCAGACAACAACAUCA--- -------------.........((((..((.((((..(---------(((..---((((((((..(((......)))..)).))))))....)))).)))).))....)))).--- ( -15.60, z-score = -1.30, R) >droMoj3.scaffold_6540 6436029 98 - 34148556 CCGCUGGAAAAAUCGC------GAUGACACGCAGCAUG---------AAAUGCCGGUAAAGAAAACAAAUUAAAUUUAAUCGAUUUGGGAAAAGCUGCCAGCCAGC-CAGUGUA-- .((((((.......((------(......)))......---------....((.((((.......(((((..(......)..)))))........)))).))...)-)))))..-- ( -20.46, z-score = 0.15, R) >droVir3.scaffold_12855 6107925 105 - 10161210 --GGUUGAAAAAUCGCCAUGACGAUGAUGGGCAGAACG---------AAAAGCCGGCAAAGAAAACAAAUUAAAUUUAAUCGAUUUGUGAAAAGCUGCCAGCCAGCGCAGACCGUC --((((....)))).....((((.((...(((.....(---------......)((((......((((((..(......)..)))))).......)))).)))....))...)))) ( -21.02, z-score = 0.24, R) >droGri2.scaffold_14906 6211032 105 + 14172833 --AGUAGAAAAAUCGCGAUGUUGAUGACGGGCAGCACG---------ACAAGACGGCAAAGAAAACAAAUUAAAUUUAAUCGAUUUGUGAAAAGCUGCCAGCCAGCACAGACGGGC --..........((((..(((((......((((((.((---------......)).........((((((..(......)..)))))).....))))))...)))))..).))).. ( -22.40, z-score = -0.55, R) >consensus _________AAGCUGCGACGAGGAUGACUGGCAGCGGA___________AAGCCGGCAAGGAAAACAAAUUAAAUUUAAUCGAUUUGUGAAAAGCUGCCAGACAACAACUACA___ .............................((((((................(....).......((((((..(......)..)))))).....))))))................. (-10.72 = -11.25 + 0.53)
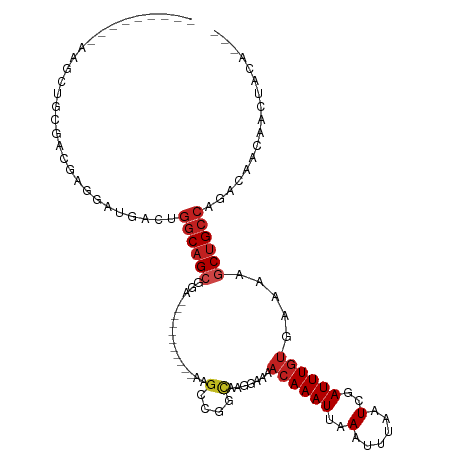
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:56:04 2011