
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 3,214,385 – 3,214,480 |
| Length | 95 |
| Max. P | 0.554909 |

| Location | 3,214,385 – 3,214,480 |
|---|---|
| Length | 95 |
| Sequences | 9 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 81.57 |
| Shannon entropy | 0.37897 |
| G+C content | 0.47180 |
| Mean single sequence MFE | -23.88 |
| Consensus MFE | -16.48 |
| Energy contribution | -16.90 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.554909 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 3214385 95 - 27905053 CGAAGGCCGCGCCAAGGCGAAACGGAAAUGAAAUCAAGCAAAUUGAAUGCGAUGAGUUGGACGUGGAAAUACGUGCCAACGGUUGUCAUAACAGC----- .((..(((.(((....)))..............(((.(((.......)))..)))((((((((((....))))).))))))))..))........----- ( -26.40, z-score = -1.26, R) >droPer1.super_6 1937598 99 - 6141320 ACAGGACAGCGCGAAAACGAAGCGGAAAUAAAAUCAAGCAAAUUGAAUGCGAUGGCUUGGACGUGGGAAUACGUGUCAACCGAUGUCAUAACAACGAGG- ....((((.(((.........))((.........(((((..((((....)))).)))))((((((......))))))..))).))))............- ( -19.90, z-score = -0.09, R) >dp4.chr2 26608860 99 - 30794189 ACAGAACAGCGCGAAAACGAAGCGGAAAUAAAAUCAAGCAAAUUGAAUGCGAUGGCUUGGACGUGGGAAUACGUGCCAACCGAUGUCAUAACAACGAGG- .........(((.........))).........((((.....))))(((((.(((.(((((((((....))))).))))))).)).)))..........- ( -18.70, z-score = 0.20, R) >droAna3.scaffold_13340 3003220 93 - 23697760 --CGAAAAAGGUGACUUGGCGUAGGAAAUAAAAUCAAGCAAAUUGAAUGCGAUGAGUUGGACGUGGGAAUACGUGCCAACCGUUGUCAUAACGGC----- --.....(((....)))(.(((...........((((.....))))((((((((.((((((((((....))))).)))))))))).))).))).)----- ( -25.90, z-score = -2.42, R) >droEre2.scaffold_4770 3473486 95 - 17746568 CGAAGGCCGAGCCAAGGCGAAACGGAAAUGAAAUCAAGCAAAUUGAAUGCGAUGAGUUGGACGUGGGAAUACGUGCCAACCGUUGUCAUAACAGC----- ......(((.((....))....)))........((((.....))))((((((((.((((((((((....))))).)))))))))).)))......----- ( -25.50, z-score = -1.36, R) >droYak2.chr3R_random 911418 95 - 3277457 CGAAGGCCAAGCCAAGGCGAAUCGGAAAUGAAAUCAAGCAAAUUGAAUGCGAUGAGUUGGACGUGGGAAUACGUGCCAACCGUUGUCAUAACAGC----- (((..(((.......)))...))).........((((.....))))((((((((.((((((((((....))))).)))))))))).)))......----- ( -25.40, z-score = -1.30, R) >droSec1.super_6 3306055 94 - 4358794 CGAAGGCCGCGCCAAGGCGAAACGGAAAUGAAAUCAA-CAAAUUGAAUGCGAUGAGUUGGACGUGGGAAUACGUGCCAACCGUUGUCAUAACAGC----- ......((((((....)))...)))........((((-....))))((((((((.((((((((((....))))).)))))))))).)))......----- ( -25.90, z-score = -1.10, R) >droSim1.chr3R 3258762 95 - 27517382 CGAAGGCCGCGCCAAGGCGAAACGGAAAUGAAAUCAAGCAAAUUGAAUGCGAUGAGUUGGACGUGGGAAUACGUGCCAACCGUUGUCAUAACAGC----- ......((((((....)))...)))........((((.....))))((((((((.((((((((((....))))).)))))))))).)))......----- ( -26.90, z-score = -1.19, R) >droWil1.scaffold_181089 7091819 88 - 12369635 ------------GCCAAGAAAUAGGAAAUAAAAUCAAGUAAAUUGAAUGCGAUGACUUGGACGUGGGAAUACGUGCCAAGCUUUGGCAUAAUAACAAGGA ------------((((((...............((((.....)))).........((((((((((....))))).))))).))))))............. ( -20.30, z-score = -2.47, R) >consensus CGAAGGCCGCGCCAAGGCGAAACGGAAAUGAAAUCAAGCAAAUUGAAUGCGAUGAGUUGGACGUGGGAAUACGUGCCAACCGUUGUCAUAACAGC_____ .................................((((.....))))((((((((.((((((((((....))))).)))))))))).)))........... (-16.48 = -16.90 + 0.42)

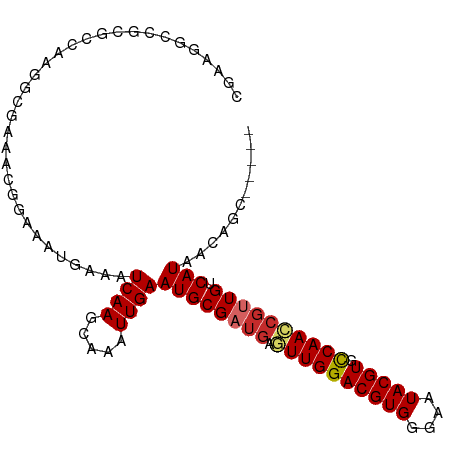
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:56:02 2011