| Sequence ID | dm3.chr3R |
|---|---|
| Location | 3,197,632 – 3,197,730 |
| Length | 98 |
| Max. P | 0.596689 |

| Location | 3,197,632 – 3,197,730 |
|---|---|
| Length | 98 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 62.37 |
| Shannon entropy | 0.71197 |
| G+C content | 0.44332 |
| Mean single sequence MFE | -29.70 |
| Consensus MFE | -5.51 |
| Energy contribution | -5.81 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.19 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.596689 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 3197632 98 + 27905053 -------GUUGCGUAUACGCAACAUAUGC---GAGAAUGAAAAAUGAAAAUAAUUGUUGAAAAACUGAUAGCUAUUCGGCUCACUCCUCUC-----AGCAGUGUUCGUCCCAU -------((((((....))))))......---...........(((((....((((((((.........((((....))))........))-----)))))).)))))..... ( -21.53, z-score = -1.43, R) >droPer1.super_6 1921434 103 + 6141320 -------GUUGCG---GCGCAAUGUGUAGCCGGAGAAUGGAAAAUGAAAAUAAUUUUUCAAAAACGGCCAGCCACGGGCUAAAAUCCCCACUGGCUGGCACUGUGCGUAUUAU -------......---(((((..((((.((((.....((((((((.......))))))))....))))((((((.(((........)))..))))))))))..)))))..... ( -37.10, z-score = -2.81, R) >dp4.chr2 26592705 103 + 30794189 -------GUUGCG---GCGCAAUGUGUAGCCGGAGAAUGGAAAAUGAAAAUAAUUUUUCAAAAACGGCCAGCCACGGGCUAAAAUCCCCACUGGCUGGCACUGUGCGUAUUAU -------......---(((((..((((.((((.....((((((((.......))))))))....))))((((((.(((........)))..))))))))))..)))))..... ( -37.10, z-score = -2.81, R) >droAna3.scaffold_13340 2988422 105 + 23697760 ---CUUUUUCAUUUAAAACCUUUUUUUCAAUUGGGCCAAAAAAUUUGAAACAUUUCUUCCAAAAUGUCCAUCCAUCGGUGAAGCGCCCGCC-----GGCACUAUGAAGUAUAU ---.....................(((((..((((((............((((((......)))))).........((((.......))))-----))).))))))))..... ( -15.90, z-score = 0.37, R) >droEre2.scaffold_4770 3456774 94 + 17746568 GCCACACGUUGCGUAUACGCAACGUAUGC---GGGAAUGAAAA-----------UUUUUAAAAACUGCCCGCUAUGCGGCUCACUCCUCUC-----AGCAGUAUUCGUGUUAU (((....((((((....))))))((((((---(((........-----------.............))))).)))))))...........-----................. ( -25.90, z-score = -1.56, R) >droSec1.super_6 3284151 105 + 4358794 GGCACGCGUUGCGUAUACGCAACGUAUGC---GAGAAUGAAAAAUGAAAAUAAUUGUUGAAAACCUGCUAGCCAUUCGGCUCACUCCUCUC-----AGGAGUGUUCGUCCUAU .(((..(((((((....)))))))..)))---.((.(((((............................((((....))))((((((....-----.))))))))))).)).. ( -35.20, z-score = -3.74, R) >droSim1.chr3R 3242233 105 + 27517382 GGCACGCGUUGCGUAUACGCAACGUAUGC---GAGAAUGAAAAAUGAAAAUAAUUGUUGAAAACCUGCUAGCCAUUCGGCUCACUCCUCUC-----AGGAGUGUUCGUCCUAU .(((..(((((((....)))))))..)))---.((.(((((............................((((....))))((((((....-----.))))))))))).)).. ( -35.20, z-score = -3.74, R) >consensus _______GUUGCGUAUACGCAACGUAUGC___GAGAAUGAAAAAUGAAAAUAAUUGUUCAAAAACUGCCAGCCAUGCGGCUCACUCCUCUC_____AGCAGUGUUCGUCCUAU .......((((((....)))))).......................................................................................... ( -5.51 = -5.81 + 0.31)

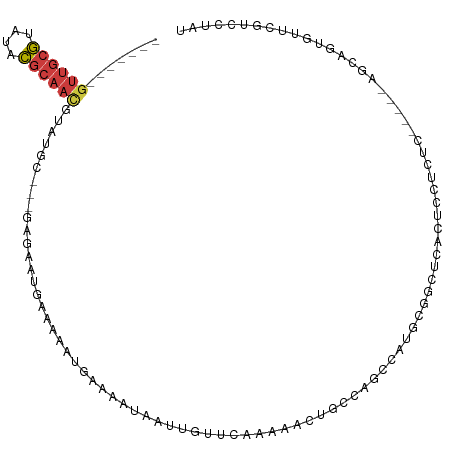

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:56:02 2011