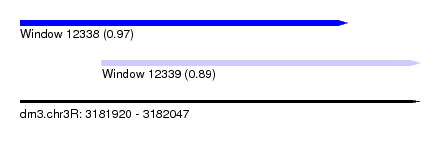
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 3,181,920 – 3,182,047 |
| Length | 127 |
| Max. P | 0.974456 |

| Location | 3,181,920 – 3,182,024 |
|---|---|
| Length | 104 |
| Sequences | 8 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 75.85 |
| Shannon entropy | 0.45092 |
| G+C content | 0.32609 |
| Mean single sequence MFE | -19.05 |
| Consensus MFE | -12.06 |
| Energy contribution | -12.19 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.63 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.974456 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
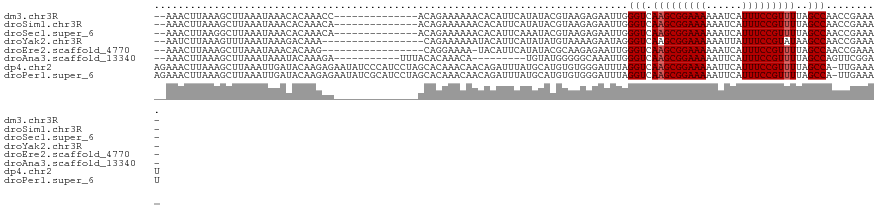
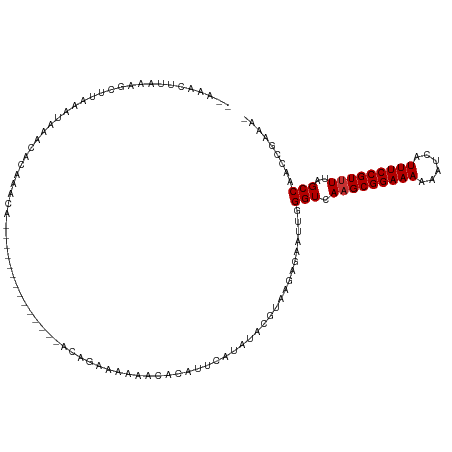
>dm3.chr3R 3181920 104 + 27905053 --AAACUUAAAGCUUAAAUAAACACAAACC--------------ACAGAAAAAACACAUUCAUAUACGUAAGAGAAUUGGGUCAAGCGGAAAAAAUCAUUUCCGUUUUAGCCAACCGAAA- --............................--------------..............(((.....(....)....((((.(.(((((((((......))))))))).).))))..))).- ( -16.20, z-score = -2.14, R) >droSim1.chr3R 3225583 104 + 27517382 --AAACUUAAAGCUUAAAUAAACACAAACA--------------ACAGAAAAAACACAUUCAUAUACGUAAGAGAAUUGGGUCAAGCGGAAAAAAUCAUUUCCGUUUUAGCCAACCGAAA- --............................--------------..............(((.....(....)....((((.(.(((((((((......))))))))).).))))..))).- ( -16.20, z-score = -2.30, R) >droSec1.super_6 3268352 104 + 4358794 --AAACUUAAGGCUUAAAUAAACACAAACA--------------ACAGAAAAAACACAUUCAAAUACGUAAGAGAAUUGGGUCAAGCGGAAAAAAUCAUUUCCGUUUUAGCCAACCGAAA- --........((..................--------------......................(....)....((((.(.(((((((((......))))))))).).))))))....- ( -16.40, z-score = -1.88, R) >droYak2.chr3R 21673474 101 - 28832112 --AAUCUUAAAGUUUAAAUAAAGACAAA-----------------CAGAAAAAAUACAUUCAUAUAUGUAAAAGAAUAGGGUCAAGCGGAAAAAAUUAUUUCCGUAUAAGCCAACCGAAA- --..((((...((((......))))...-----------------.........(((((......))))).))))....(((...(((((((......)))))))....)))........- ( -16.40, z-score = -2.92, R) >droEre2.scaffold_4770 3442260 100 + 17746568 --AAACUUAAAGCUUAAAUAAACACAAG-----------------CAGGAAAA-UACAUUCAUAUACGCAAGAGAAUUGGGUCAAGCGGAAAAAAUCAUUUCCGUUUUAGCCAACCGAAA- --.........((((..........)))-----------------).((....-............(....)....((((.(.(((((((((......))))))))).).))))))....- ( -20.20, z-score = -3.08, R) >droAna3.scaffold_13340 2973038 98 + 23697760 --AAACUUAAAGCUUAAAUAAAUACAAAGA-----------UUUACACAAACA---------UGUAUGGGGGCAAAUUGGGUCAAGCGGAAAAAUUCAUUUCCGUUUUAGCCAGUUCGGA- --.........((((......(((((....-----------............---------)))))..)))).((((((.(.(((((((((......))))))))).).))))))....- ( -20.69, z-score = -1.95, R) >dp4.chr2 26580948 120 + 30794189 AGAAACUUAAAGCUUAAAUUGAUACAAGAGAAUAUCCCAUCCUAGCACAAACAACAGAUUUAUGCAUGUGUGGGAUUUAGGUCAAGCGGAAAAAUUCAUUUCCGUUUUAGCCA-UUGAAAU ........................(((......((((((..(..(((.((((....).))).)))..)..))))))...(((.(((((((((......)))))))))..))).-))).... ( -24.10, z-score = -1.29, R) >droPer1.super_6 1909742 120 + 6141320 AGAAACUUAAAGCUUAAAUUGAUACAAGAGAAUAUCGCAUCCUAGCACAAACAACAGAUUUAUGCAUGUGUGGGAUUUAGGUCAAGCGGAAAAAUUCAUUUCCGUUUUAGCCA-UUGAAAU ....................((((........))))..((((((.((((..((.........))..))))))))))...(((.(((((((((......)))))))))..))).-....... ( -22.20, z-score = -0.53, R) >consensus __AAACUUAAAGCUUAAAUAAACACAAACA______________ACAGAAAAAACACAUUCAUAUACGUAAGAGAAUUGGGUCAAGCGGAAAAAAUCAUUUCCGUUUUAGCCAACCGAAA_ ...............................................................................(((.(((((((((......)))))))))..)))......... (-12.06 = -12.19 + 0.12)
| Location | 3,181,946 – 3,182,047 |
|---|---|
| Length | 101 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.21 |
| Shannon entropy | 0.53440 |
| G+C content | 0.41847 |
| Mean single sequence MFE | -23.11 |
| Consensus MFE | -12.06 |
| Energy contribution | -12.19 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.893332 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 3181946 101 + 27905053 --CCACAGAAAAAACACAUUCAUAUACGUAAGAGAAUUGGGUCAAGCGGAAAAAAUCAUUUCCGUUUUAGCCAACCGAAAAGGGUGCCCAAAACAGAUGUGAC----------------- --............((((((......(....)....((((((.(((((((((......)))))))))......(((......)))))))))....))))))..----------------- ( -24.70, z-score = -2.85, R) >droSim1.chr3R 3225609 101 + 27517382 --CAACAGAAAAAACACAUUCAUAUACGUAAGAGAAUUGGGUCAAGCGGAAAAAAUCAUUUCCGUUUUAGCCAACCGAAAAGGGUGCCCAAAACAGAUGGCAC----------------- --........................(....)....((((.(.(((((((((......))))))))).).))))((.....))(((((..........)))))----------------- ( -21.90, z-score = -2.09, R) >droSec1.super_6 3268378 101 + 4358794 --CAACAGAAAAAACACAUUCAAAUACGUAAGAGAAUUGGGUCAAGCGGAAAAAAUCAUUUCCGUUUUAGCCAACCGAAAAGGGUCCCCAAAACAGAUGGGAC----------------- --........................(....)....((((.(.(((((((((......))))))))).).))))((.....))(((((..........)))))----------------- ( -22.10, z-score = -1.87, R) >droYak2.chr3R 21673500 98 - 28832112 -----CAGAAAAAAUACAUUCAUAUAUGUAAAAGAAUAGGGUCAAGCGGAAAAAAUUAUUUCCGUAUAAGCCAACCGAAAAGGGUGCCCAAAACAUAUGGGAC----------------- -----.........(((((......))))).........(((...(((((((......)))))))....))).(((......))).((((.......))))..----------------- ( -20.00, z-score = -2.20, R) >droEre2.scaffold_4770 3442286 97 + 17746568 -----CAGGAAAA-UACAUUCAUAUACGCAAGAGAAUUGGGUCAAGCGGAAAAAAUCAUUUCCGUUUUAGCCAACCGAAAAGGGUGCCCAAAACAGAUGGCAC----------------- -----..((....-............(....)....((((.(.(((((((((......))))))))).).)))))).......(((((..........)))))----------------- ( -23.40, z-score = -2.30, R) >droAna3.scaffold_13340 2973064 112 + 23697760 --------GAUUUACACAAACAUGUAUGGGGGCAAAUUGGGUCAAGCGGAAAAAUUCAUUUCCGUUUUAGCCAGUUCGGAAGGGUGGAAGUUGAAAUUGGCUGGGAACAAAGUUUGCCAC --------....((((......))))....((((((((..((((((((((((......)))))))))((((((((((((...........))).))))))))).).))..)))))))).. ( -31.00, z-score = -1.82, R) >dp4.chr2 26580988 103 + 30794189 CCUAGCACAAACAACAGAUUUAUGCAUGUGUGGGAUUUAGGUCAAGCGGAAAAAUUCAUUUCCGUUUUAGCCA--UUGAAAUAGUGGACGCGGAGGAGGAGGGGC--------------- (((.(((.((((....).))).))).(((((..(((....)))(((((((((......)))))))))...(((--(((...))))))))))).......)))...--------------- ( -20.90, z-score = 0.38, R) >droPer1.super_6 1909782 103 + 6141320 CCUAGCACAAACAACAGAUUUAUGCAUGUGUGGGAUUUAGGUCAAGCGGAAAAAUUCAUUUCCGUUUUAGCCA--UUGAAAUAGUGGACGCGGAGGAGGAGGGGC--------------- (((.(((.((((....).))).))).(((((..(((....)))(((((((((......)))))))))...(((--(((...))))))))))).......)))...--------------- ( -20.90, z-score = 0.38, R) >consensus ___A_CAGAAAAAACACAUUCAUAUACGUAAGAGAAUUGGGUCAAGCGGAAAAAAUCAUUUCCGUUUUAGCCAACCGAAAAGGGUGCCCAAAACAGAUGGGAC_________________ .......................................(((.(((((((((......)))))))))..)))................................................ (-12.06 = -12.19 + 0.12)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:56:00 2011