
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 3,181,591 – 3,181,688 |
| Length | 97 |
| Max. P | 0.824192 |

| Location | 3,181,591 – 3,181,688 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 95.60 |
| Shannon entropy | 0.08201 |
| G+C content | 0.45361 |
| Mean single sequence MFE | -28.53 |
| Consensus MFE | -22.17 |
| Energy contribution | -23.00 |
| Covariance contribution | 0.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.34 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.734534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 3181591 97 + 27905053 GGCGUGAGCCAUAAAGCUAACAAAUUGCUGACACAACAUAUGAAUGACAAUAUAUCAGUGGCGGAACCAGCCAGCUGAGUGGCUUCUGCUGAAUAAC ((((.(((((((..((((........(((((......((((........)))).)))))(((.......)))))))..))))))).))))....... ( -28.20, z-score = -2.23, R) >droSim1.chr3R 3225254 97 + 27517382 GGCGUGAGCCAUAAAGCUAACAAAUUGCUGACACAACAUAUGAAUGACAAUAUAUCAGUGGCGGAACCAGCCAGCUGAGUGGCUGCUGCUGAAUAAC (((....)))....(((.........))).........................((((..((((.(((((....))).))..))))..))))..... ( -29.60, z-score = -2.45, R) >droSec1.super_6 3268023 97 + 4358794 GGCGUGAGCCAUAAAGCUAACAAAUUGCUGACACAACAUAUGAAUGACAAUAUAUCAGUGGCGGAACCAGCCAGCUGAGUGGCUGCUGCUGAAUAAC (((....)))....(((.........))).........................((((..((((.(((((....))).))..))))..))))..... ( -29.60, z-score = -2.45, R) >droYak2.chr3R 21673148 97 - 28832112 GGCUUCAGCCAUAAAGCUAACAAAUUGCUGACACAACAUAUGAAUGACAAUAUAUCAGUGGCGAAACCAGCCAGCUAAGUGGCUGCUGCUGAAAAAC (((..(((((((..((((........(((((......((((........)))).)))))(((.......)))))))..)))))))..)))....... ( -27.90, z-score = -2.68, R) >droEre2.scaffold_4770 3441933 97 + 17746568 GGCGCGAGCCAUAAAGCUAACAAAUUGCUGACACAACAUAUGAAUGACAAUAUAUCAGUGGCGGAACCAGCCAGCUGAGUGGCUGCUGCUGAAUAAC (((....)))....(((.........))).........................((((..((((.(((((....))).))..))))..))))..... ( -29.10, z-score = -2.24, R) >droAna3.scaffold_13340 2972711 97 + 23697760 GGCUUCAGCCAUAAAGCUAACAAAUUGCUGACACAACAUAUGAAUGACAAUAUAUCAGUUGCGGAACAAGGCAGCUGAGUGGCUGCUGGUGAAAAAC .(((.(((((((..(((.........))).........................((((((((........)))))))))))))))..)))....... ( -26.80, z-score = -1.98, R) >consensus GGCGUGAGCCAUAAAGCUAACAAAUUGCUGACACAACAUAUGAAUGACAAUAUAUCAGUGGCGGAACCAGCCAGCUGAGUGGCUGCUGCUGAAUAAC ((((..((((((..((((........(((((......((((........)))).)))))(((.......)))))))..))))))..))))....... (-22.17 = -23.00 + 0.83)



| Location | 3,181,591 – 3,181,688 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 95.60 |
| Shannon entropy | 0.08201 |
| G+C content | 0.45361 |
| Mean single sequence MFE | -28.20 |
| Consensus MFE | -25.94 |
| Energy contribution | -26.00 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.824192 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 3181591 97 - 27905053 GUUAUUCAGCAGAAGCCACUCAGCUGGCUGGUUCCGCCACUGAUAUAUUGUCAUUCAUAUGUUGUGUCAGCAAUUUGUUAGCUUUAUGGCUCACGCC ........((...(((((...((((((((((.....)))((((((((....(((....))).))))))))......)))))))...)))))...)). ( -27.60, z-score = -1.55, R) >droSim1.chr3R 3225254 97 - 27517382 GUUAUUCAGCAGCAGCCACUCAGCUGGCUGGUUCCGCCACUGAUAUAUUGUCAUUCAUAUGUUGUGUCAGCAAUUUGUUAGCUUUAUGGCUCACGCC ........((...(((((...((((((((((.....)))((((((((....(((....))).))))))))......)))))))...)))))...)). ( -27.60, z-score = -1.38, R) >droSec1.super_6 3268023 97 - 4358794 GUUAUUCAGCAGCAGCCACUCAGCUGGCUGGUUCCGCCACUGAUAUAUUGUCAUUCAUAUGUUGUGUCAGCAAUUUGUUAGCUUUAUGGCUCACGCC ........((...(((((...((((((((((.....)))((((((((....(((....))).))))))))......)))))))...)))))...)). ( -27.60, z-score = -1.38, R) >droYak2.chr3R 21673148 97 + 28832112 GUUUUUCAGCAGCAGCCACUUAGCUGGCUGGUUUCGCCACUGAUAUAUUGUCAUUCAUAUGUUGUGUCAGCAAUUUGUUAGCUUUAUGGCUGAAGCC ........((..((((((...((((((((((.....)))((((((((....(((....))).))))))))......)))))))...))))))..)). ( -32.70, z-score = -2.88, R) >droEre2.scaffold_4770 3441933 97 - 17746568 GUUAUUCAGCAGCAGCCACUCAGCUGGCUGGUUCCGCCACUGAUAUAUUGUCAUUCAUAUGUUGUGUCAGCAAUUUGUUAGCUUUAUGGCUCGCGCC ........((.(((((((...((((((((((.....)))((((((((....(((....))).))))))))......)))))))...))))).)))). ( -30.60, z-score = -2.06, R) >droAna3.scaffold_13340 2972711 97 - 23697760 GUUUUUCACCAGCAGCCACUCAGCUGCCUUGUUCCGCAACUGAUAUAUUGUCAUUCAUAUGUUGUGUCAGCAAUUUGUUAGCUUUAUGGCUGAAGCC ...........(((((......)))))........((..((((((((....(((....))).)))))))).......((((((....)))))).)). ( -23.10, z-score = -0.92, R) >consensus GUUAUUCAGCAGCAGCCACUCAGCUGGCUGGUUCCGCCACUGAUAUAUUGUCAUUCAUAUGUUGUGUCAGCAAUUUGUUAGCUUUAUGGCUCACGCC ........((...(((((...((((((((((.....)))((((((((....(((....))).))))))))......)))))))...)))))...)). (-25.94 = -26.00 + 0.06)

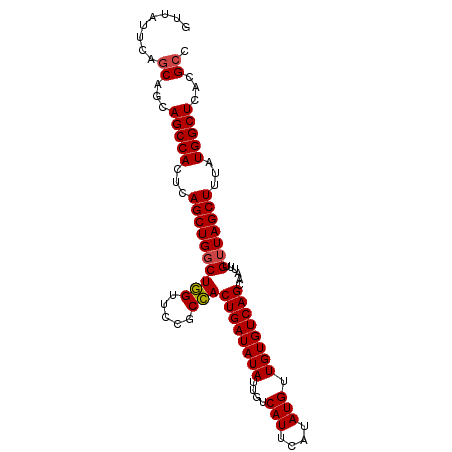
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:55:58 2011