
| Sequence ID | dm3.chr3R |
|---|---|
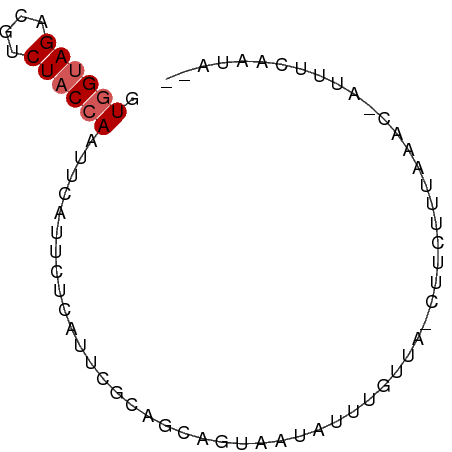
| Location | 3,166,003 – 3,166,063 |
| Length | 60 |
| Max. P | 0.830710 |

| Location | 3,166,003 – 3,166,063 |
|---|---|
| Length | 60 |
| Sequences | 5 |
| Columns | 74 |
| Reading direction | forward |
| Mean pairwise identity | 77.89 |
| Shannon entropy | 0.38867 |
| G+C content | 0.33725 |
| Mean single sequence MFE | -10.22 |
| Consensus MFE | -5.82 |
| Energy contribution | -6.22 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.705274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
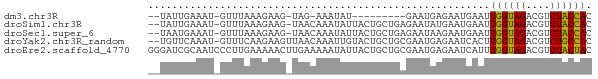
>dm3.chr3R 3166003 60 + 27905053 GUGGUAGACGUCUACCAAUUCAUUCUCAUUC---------AUAUUU-CUA-CUUCUUUAAAC-AUUUCAAUA-- .((((((....))))))..............---------......-...-...........-.........-- ( -8.00, z-score = -3.57, R) >droSim1.chr3R 3215339 70 + 27517382 GUGGUAGACGUCUACCAAUUCAUUCAUAUUCUCAGCAGUAAUAUUUGUUA-CUUCUUUAAAC-AUUUCAAUA-- .((((((....))))))................((.((((((....))))-)).))......-.........-- ( -11.70, z-score = -3.22, R) >droSec1.super_6 3258106 70 + 4358794 GUGGUAGACGUCUACCAAUUCAUUCUUAUUCUCAGCAGUAAUAUUUGUUA-CUUCUUUAAAC-AUUUCAUUA-- .((((((....))))))................((.((((((....))))-)).))......-.........-- ( -11.70, z-score = -3.24, R) >droYak2.chr3R_random 885692 71 + 3277457 GUGGCAGACGUCUACCAAGUGAUUCUCAUUCGCAGCAGUACAAUUUGUUAACUUCUUGAAAC-AUUUGAACA-- (((((....).))))((((((.(((........(((((......)))))........))).)-)))))....-- ( -10.79, z-score = 0.55, R) >droEre2.scaffold_4770 3432116 74 + 17746568 GUAGUAGACGUCUACCAAAUGAUUCUCAUUCGCAGCAGUAAUAUUUUUCAAGUUUUUCAAGGGAUUGCGAUCCC ((.((((....))))..((((.....)))).))...........................(((((....))))) ( -8.90, z-score = 0.65, R) >consensus GUGGUAGACGUCUACCAAUUCAUUCUCAUUCGCAGCAGUAAUAUUUGUUA_CUUCUUUAAAC_AUUUCAAUA__ .((((((....))))))......................................................... ( -5.82 = -6.22 + 0.40)


| Location | 3,166,003 – 3,166,063 |
|---|---|
| Length | 60 |
| Sequences | 5 |
| Columns | 74 |
| Reading direction | reverse |
| Mean pairwise identity | 77.89 |
| Shannon entropy | 0.38867 |
| G+C content | 0.33725 |
| Mean single sequence MFE | -12.40 |
| Consensus MFE | -7.56 |
| Energy contribution | -7.24 |
| Covariance contribution | -0.32 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.830710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 3166003 60 - 27905053 --UAUUGAAAU-GUUUAAAGAAG-UAG-AAAUAU---------GAAUGAGAAUGAAUUGGUAGACGUCUACCAC --.........-((((..(...(-((.-...)))---------...)..))))....((((((....)))))). ( -9.00, z-score = -2.03, R) >droSim1.chr3R 3215339 70 - 27517382 --UAUUGAAAU-GUUUAAAGAAG-UAACAAAUAUUACUGCUGAGAAUAUGAAUGAAUUGGUAGACGUCUACCAC --.......((-((((..((.((-(((......))))).))..))))))........((((((....)))))). ( -14.40, z-score = -3.19, R) >droSec1.super_6 3258106 70 - 4358794 --UAAUGAAAU-GUUUAAAGAAG-UAACAAAUAUUACUGCUGAGAAUAAGAAUGAAUUGGUAGACGUCUACCAC --........(-((((..((.((-(((......))))).))..))))).........((((((....)))))). ( -13.40, z-score = -3.04, R) >droYak2.chr3R_random 885692 71 - 3277457 --UGUUCAAAU-GUUUCAAGAAGUUAACAAAUUGUACUGCUGCGAAUGAGAAUCACUUGGUAGACGUCUGCCAC --.........-.(((((.(.(((..((.....))...))).)...)))))......((((((....)))))). ( -11.20, z-score = 0.75, R) >droEre2.scaffold_4770 3432116 74 - 17746568 GGGAUCGCAAUCCCUUGAAAAACUUGAAAAAUAUUACUGCUGCGAAUGAGAAUCAUUUGGUAGACGUCUACUAC (((((....)))))............................((((((.....))))))((((....))))... ( -14.00, z-score = -1.55, R) >consensus __UAUUGAAAU_GUUUAAAGAAG_UAACAAAUAUUACUGCUGAGAAUGAGAAUGAAUUGGUAGACGUCUACCAC .........................................................((((((....)))))). ( -7.56 = -7.24 + -0.32)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:55:55 2011