
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 3,161,271 – 3,161,367 |
| Length | 96 |
| Max. P | 0.869955 |

| Location | 3,161,271 – 3,161,367 |
|---|---|
| Length | 96 |
| Sequences | 9 |
| Columns | 97 |
| Reading direction | forward |
| Mean pairwise identity | 73.46 |
| Shannon entropy | 0.53025 |
| G+C content | 0.47806 |
| Mean single sequence MFE | -26.06 |
| Consensus MFE | -9.11 |
| Energy contribution | -9.42 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.519863 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 3161271 96 + 27905053 GACAGUAGCGCCUCCAGUGAAAUAAAUCACAGAAAUA-UGUCUUUCAUCUCGGCGCCUGCAGCUUGCAGCUUUGCAGUUGCAAGUUGGCUGUCUUCG ((((((.(((((....((((......)))).((((..-....)))).....)))))...((((((((((((....)))))))))))))))))).... ( -40.40, z-score = -5.02, R) >droPer1.super_6 1897163 86 + 6141320 GACAGUAGCGCCUCCAGUGGUAUAAAUCACAGAAAUA-UGUCUUUCAUCUUGGCGCUUGCAC----------UUCAGUCUCUGGUCUCCAGCCUCCA ((((((((((((....(((((....))))).((((..-....)))).....))))))...))----------)...))).((((...))))...... ( -20.70, z-score = -1.28, R) >dp4.chr2 26568335 79 + 30794189 GACAGUAGCGCCUCCAGUGAUAUAAAUCACAGAAAUA-UGUCUUUCAUCUUGGCGCUUGCAC----------UUCAGUCUCUGGUCUCCA------- (((.((((((((....(((((....))))).((((..-....)))).....))))).)))..----------..(((...))))))....------- ( -21.20, z-score = -2.37, R) >droEre2.scaffold_4770 3427301 81 + 17746568 GACAGUAGCGCCUCCAGUGAAAUAAAUCACAGAAAUA-UGUCUUUCAUCUCGGCGCCUGCAG--------UUUGCAGUUGCGAGUUGGCA------- ..(((..(((((....((((......)))).((((..-....)))).....))))))))((.--------.((((....))))..))...------- ( -22.20, z-score = -0.80, R) >droSec1.super_6 3253177 96 + 4358794 GACAGUAGCGCCUCCAGUGAAAUAAAUCACAGAAAUA-UGUCUUUCAUCUCGGCGCCUGCAGUUUGCAACUUUGCAGUUGCGAGUUGGCUGUCUUCA ((((((.(((((....((((......)))).((((..-....)))).....)))))...((..((((((((....))))))))..)))))))).... ( -35.00, z-score = -3.68, R) >droSim1.chr3R 3209862 96 + 27517382 GACAGUAGCGCCUCCAGUGAAAUAAAUCACAGAAAUA-UGUCUUUCAUCUCGGCGCCUGCAGUUUGCAGCUUUGCAGUUGCGAGUUGGCUGUCUUCA ((((((.(((((....((((......)))).((((..-....)))).....)))))...((..((((((((....))))))))..)))))))).... ( -35.00, z-score = -3.20, R) >droMoj3.scaffold_6540 27817569 85 - 34148556 GACAGUAGCGCCUGCAGAAAAAUAAAUCACAGAAAAA-UGUCUUUAAUCUUGGCGCCUCGAG------AGGCGGCGGUA-CAGGUCGUCCGCG---- (((.(((.((((.((.......((((..(((......-))).))))......))((((....------)))))))).))-)..))).......---- ( -23.52, z-score = -0.62, R) >droVir3.scaffold_12855 4146087 82 - 10161210 GACAGUAGCGCCUGCAGUAAAAUAAAUCACAGAAAAA-UGUCUUUAAUCUUGGCGCCU---G------AAGUUGCGGUA-CAGGUCAGUCGCA---- ..(((..(((((...((.....((((..(((......-))).))))..)).)))))))---)------....(((((..-........)))))---- ( -13.90, z-score = 1.50, R) >droGri2.scaffold_14906 8041763 85 + 14172833 --CGGUAGCGCCUGAAGUUAAAUAAAUCUCAAAAAAAAUGUCUUUAAUCUUGGCGCCUU---------GCGUUGCUGUU-CAGGUCGCCGCCGACCA --(((..(((((((((((((((....................))))))...(((((...---------)))))....))-)))).)))..))).... ( -22.65, z-score = -1.57, R) >consensus GACAGUAGCGCCUCCAGUGAAAUAAAUCACAGAAAUA_UGUCUUUCAUCUUGGCGCCUGCAG_______CUUUGCAGUUGCAGGUCGGCAGCC__C_ .......(((((....((((((....................))))))...)))))......................................... ( -9.11 = -9.42 + 0.31)

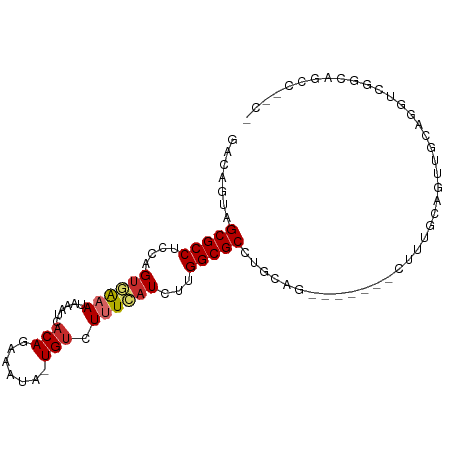
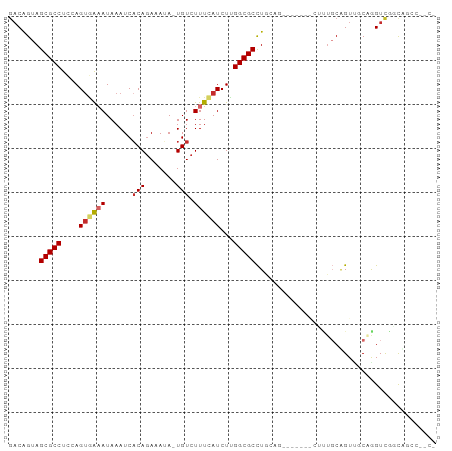
| Location | 3,161,271 – 3,161,367 |
|---|---|
| Length | 96 |
| Sequences | 9 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 73.46 |
| Shannon entropy | 0.53025 |
| G+C content | 0.47806 |
| Mean single sequence MFE | -24.46 |
| Consensus MFE | -12.92 |
| Energy contribution | -12.94 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.869955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3R 3161271 96 - 27905053 CGAAGACAGCCAACUUGCAACUGCAAAGCUGCAAGCUGCAGGCGCCGAGAUGAAAGACA-UAUUUCUGUGAUUUAUUUCACUGGAGGCGCUACUGUC ....(((((.((.((((((.((....)).)))))).))..((((((((((((.......-)))))).((((......))))....)))))).))))) ( -32.20, z-score = -2.24, R) >droPer1.super_6 1897163 86 - 6141320 UGGAGGCUGGAGACCAGAGACUGAA----------GUGCAAGCGCCAAGAUGAAAGACA-UAUUUCUGUGAUUUAUACCACUGGAGGCGCUACUGUC ...((.((((...))))...))..(----------((...((((((.....((((....-..)))).(((........)))....)))))))))... ( -19.00, z-score = 0.21, R) >dp4.chr2 26568335 79 - 30794189 -------UGGAGACCAGAGACUGAA----------GUGCAAGCGCCAAGAUGAAAGACA-UAUUUCUGUGAUUUAUAUCACUGGAGGCGCUACUGUC -------.(....)(((..((....----------))...((((((.....((((....-..)))).(((((....)))))....)))))).))).. ( -19.30, z-score = -1.01, R) >droEre2.scaffold_4770 3427301 81 - 17746568 -------UGCCAACUCGCAACUGCAAA--------CUGCAGGCGCCGAGAUGAAAGACA-UAUUUCUGUGAUUUAUUUCACUGGAGGCGCUACUGUC -------(((......))).((((...--------..))))(((((((((((.......-)))))).((((......))))....)))))....... ( -23.30, z-score = -1.72, R) >droSec1.super_6 3253177 96 - 4358794 UGAAGACAGCCAACUCGCAACUGCAAAGUUGCAAACUGCAGGCGCCGAGAUGAAAGACA-UAUUUCUGUGAUUUAUUUCACUGGAGGCGCUACUGUC ....(((((.......((((((....))))))........((((((((((((.......-)))))).((((......))))....)))))).))))) ( -31.60, z-score = -2.75, R) >droSim1.chr3R 3209862 96 - 27517382 UGAAGACAGCCAACUCGCAACUGCAAAGCUGCAAACUGCAGGCGCCGAGAUGAAAGACA-UAUUUCUGUGAUUUAUUUCACUGGAGGCGCUACUGUC ....(((((.......(((..((((....))))...))).((((((((((((.......-)))))).((((......))))....)))))).))))) ( -29.40, z-score = -1.96, R) >droMoj3.scaffold_6540 27817569 85 + 34148556 ----CGCGGACGACCUG-UACCGCCGCCU------CUCGAGGCGCCAAGAUUAAAGACA-UUUUUCUGUGAUUUAUUUUUCUGCAGGCGCUACUGUC ----.((((.((.((((-((..(.(((((------....))))).).((((((.(((..-....))).)))))).......))))))))...)))). ( -23.50, z-score = -1.10, R) >droVir3.scaffold_12855 4146087 82 + 10161210 ----UGCGACUGACCUG-UACCGCAACUU------C---AGGCGCCAAGAUUAAAGACA-UUUUUCUGUGAUUUAUUUUACUGCAGGCGCUACUGUC ----((((((......)-)..))))....------.---.((((((........(((..-....)))((((......))))....))))))...... ( -17.30, z-score = -0.39, R) >droGri2.scaffold_14906 8041763 85 - 14172833 UGGUCGGCGGCGACCUG-AACAGCAACGC---------AAGGCGCCAAGAUUAAAGACAUUUUUUUUGAGAUUUAUUUAACUUCAGGCGCUACCG-- ....(((..(((.((((-((..((..(..---------..)..)).....(((((...(((((....)))))...))))).)))))))))..)))-- ( -24.50, z-score = -2.24, R) >consensus _G__GGCAGCCGACCUGCAACUGCAAAG_______CUGCAGGCGCCAAGAUGAAAGACA_UAUUUCUGUGAUUUAUUUCACUGGAGGCGCUACUGUC ........................................((((((........(((.......)))((((......))))....))))))...... (-12.92 = -12.94 + 0.03)


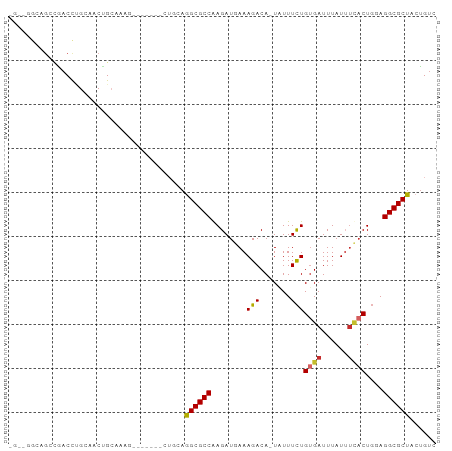
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:55:53 2011