| Sequence ID | dm3.chr2L |
|---|---|
| Location | 6,579,044 – 6,579,249 |
| Length | 205 |
| Max. P | 0.874344 |

| Location | 6,579,044 – 6,579,107 |
|---|---|
| Length | 63 |
| Sequences | 4 |
| Columns | 63 |
| Reading direction | reverse |
| Mean pairwise identity | 78.47 |
| Shannon entropy | 0.34590 |
| G+C content | 0.48114 |
| Mean single sequence MFE | -13.68 |
| Consensus MFE | -12.65 |
| Energy contribution | -11.90 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.36 |
| Mean z-score | -0.91 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.809805 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 6579044 63 - 23011544 CAAACCUUUUUGUUUUUUUUUGCGUGUAUGCCAUGAGUGCCCCUUAGUGGCAGUUCUAUGUGG ...................(..((((..(((((((((.....)))).)))))....))))..) ( -12.10, z-score = -0.74, R) >droYak2.chr2L 15987253 54 + 22324452 ---------GGUACUUUCUCUGCGUGUAUGCGAUGAGUGCCACUCGGUGGCAGUGCCAUGUGG ---------(((((...(((..(((....)))..)))((((((...)))))))))))...... ( -19.20, z-score = -1.17, R) >droSec1.super_3 2118677 59 - 7220098 GAAACC----UUUUUUUUUUUGCGUGUAUGCCGUGAGUGCCCCUUAGUGGCAGUUCUAUGUGG ......----.........(..((((..(((((((((.....)))).)))))....))))..) ( -11.70, z-score = -0.89, R) >droSim1.chr2L 6371264 56 - 22036055 GAAAC-------UUUUUUUUUGCGUGUAUGCCGUGAGUGCCCCUUAGUGGCAGUUCUAUGUGG .....-------.......(..((((..(((((((((.....)))).)))))....))))..) ( -11.70, z-score = -0.86, R) >consensus GAAAC_____UUUUUUUUUUUGCGUGUAUGCCAUGAGUGCCCCUUAGUGGCAGUUCUAUGUGG ...................(..((((..(((((((((.....)))).)))))....))))..) (-12.65 = -11.90 + -0.75)
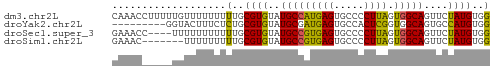
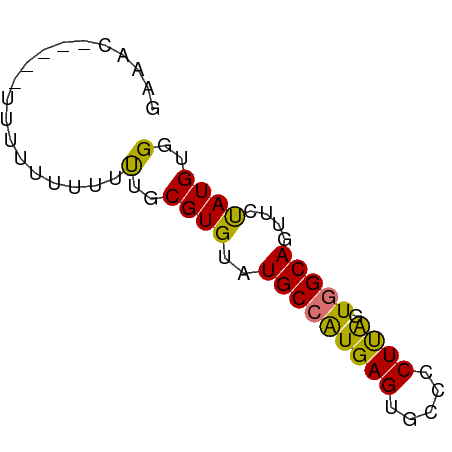
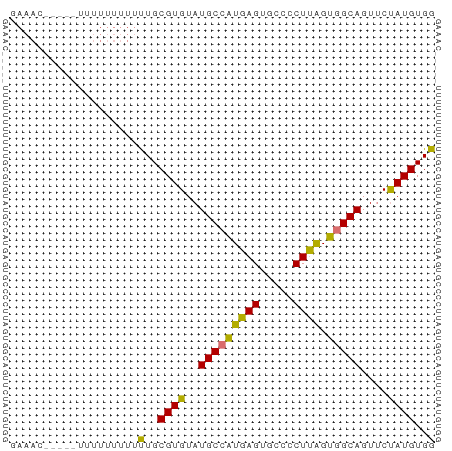
| Location | 6,579,107 – 6,579,249 |
|---|---|
| Length | 142 |
| Sequences | 5 |
| Columns | 152 |
| Reading direction | reverse |
| Mean pairwise identity | 70.17 |
| Shannon entropy | 0.53341 |
| G+C content | 0.35307 |
| Mean single sequence MFE | -35.92 |
| Consensus MFE | -11.42 |
| Energy contribution | -16.38 |
| Covariance contribution | 4.96 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.874344 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 6579107 142 - 23011544 GUGAAAAUCCACAGCAAAGGUGCUUAAAAGUAUGCAACAGCAAGCUGUAAAAGAACGUUACCAUUUCAUAGCAUUCUUUUGGGCACAAGUGGUUUUAGUAUCGAAUU----CAUAUAUGUCUUU------CGUUUUCUCUAUCAGAUUAUUA (((......))).......(((((((((((.((((.((((....))))....(((.........)))...)))).))))))))))).(((((((((((...((((..----((....))...))------))......)))..)))))))). ( -30.70, z-score = -1.63, R) >droEre2.scaffold_4929 15487223 152 - 26641161 GUGAAAAUGCCCAGUGAAGAUGCUUAAGAGUAUGCAACAACAAGCUGCAAAAGAACGUUACCUUUUCGUUGCAUACUUUUGGGCACAAGUGGAUUUAGUAUCAAAUUGGUAAAUGUAUGCUUCAGUUGCGCGUUGAUUUUAUCAGAGUAUUA .....(((((.((((((((.((((((((((((((((((...(((..((........))...)))...))))))))))))))))))...(((.(((((.((......)).))))).))).))))).))).))))).................. ( -42.20, z-score = -1.95, R) >droYak2.chr2L 15987307 152 + 22324452 CUGAAAAUCCCCAGCACAGGUGCUUAAAAGUAUGCAACAGCAAAGUGGAAAAGAACGUUACCCGUUCAUUGCAUACUUUUGGGCGCAAGUGGAUUUAGUAUCAAAUUGAUAUAUGUACGCAUCAGUUGUGUGCUCAUUUUAUCAGAUUAUUA ((((((((((.....((..((((((((((((((((((((......)).....(((((.....))))).))))))))))))))))))..)))))))).(((((.....)))))..(((((((.....)))))))........))))....... ( -49.50, z-score = -4.50, R) >droSec1.super_3 2118736 129 - 7220098 GUGAAAAUCCACAGGAAAGGUGCUUAAAAGUAUGCAACAGCAAGCUGUAAAAGAACGUUACCUUUUCAUUGCAUACUUUUGGGCACAAGUGGUUUUAGUAUCAAAUU---AGAAAUCAGAAUCA------UAUUAUUA-------------- (((......))).......(((((((((((((((((((((....))))(((((........)))))...)))))))))))))))))..((((((((.((.((.....---.)).)).)))))))------).......-------------- ( -38.40, z-score = -4.88, R) >droSim1.chr2L 6371320 112 - 22036055 GUGAAAAUCCAUAGCAAAGGUGCUUAAAAGUAUGCAACAGCAAGCUGUAAAAGAACAUUACC------------------------------UUGUAGUAUCGAAUU----UAUAUGUGCCUUG------UGUUUUCUCUAUCAGAUUAUUA (.(((((..((..((((((((((((...(((.(((....))).)))....))).....))))------------------------------))((((........)----)))...)))..))------..))))).)............. ( -18.80, z-score = 0.31, R) >consensus GUGAAAAUCCACAGCAAAGGUGCUUAAAAGUAUGCAACAGCAAGCUGUAAAAGAACGUUACCUUUUCAUUGCAUACUUUUGGGCACAAGUGGUUUUAGUAUCAAAUU___AUAUAUAUGCAUCA______UGUUUUUUCUAUCAGAUUAUUA ...................(((((((((((((((((((((....))).....(((.........))).)))))))))))))))))).................................................................. (-11.42 = -16.38 + 4.96)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:21:37 2011