
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 3,083,108 – 3,083,238 |
| Length | 130 |
| Max. P | 0.556067 |

| Location | 3,083,108 – 3,083,238 |
|---|---|
| Length | 130 |
| Sequences | 13 |
| Columns | 137 |
| Reading direction | reverse |
| Mean pairwise identity | 71.36 |
| Shannon entropy | 0.61425 |
| G+C content | 0.53442 |
| Mean single sequence MFE | -36.61 |
| Consensus MFE | -22.43 |
| Energy contribution | -22.43 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.58 |
| Mean z-score | -0.33 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.556067 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
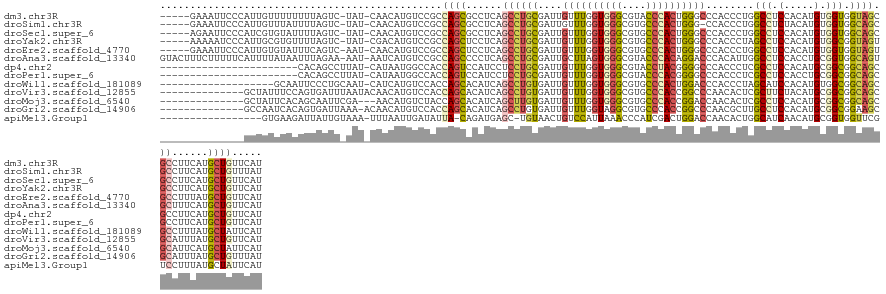
>dm3.chr3R 3083108 130 - 27905053 -----GAAAUUCCCAUUGUUUUUUUUAGUC-UAU-CAACAUGUCCGCCAGCGCCUCAGCCUGCGAUUGUUUGGUGGGCGUACCCACUGGGCCCACCCUGGCCUCCACAUGUGGUGGUAGCGCCUUCAUGCUGUUCAU -----(((.....................(-(((-((.(((((..((((((((........))).......(((((((.((.....)).))))))))))))....)))))))))))(((((......)))))))).. ( -39.10, z-score = -0.45, R) >droSim1.chr3R 3128384 129 - 27517382 -----GAAAUUCCCAUUGUUUAUUUUAGUC-UAU-CAACAUGUCCGCCAGCGCCUCAGCCUGCGAUUGUUUGGUGGGCGUGCCCACUGGG-CCACCCUGGCCUCUACAUGUGGUGGCAGCGCCUUCAUGCUGUUUAU -----........................(-(((-((.(((((..((((((((........)))...(((..(((((....)))))..))-)....)))))....)))))))))))(((((......)))))..... ( -39.60, z-score = -0.10, R) >droSec1.super_6 3175155 130 - 4358794 -----AGAAUUCCCAUCGUGUAUUUUAGUC-UAU-CAACAUGUCCGCCAGCGCCUCAGCCUGCGAUUGUUUGGUGGGCGUGCCCACUGGGCCCACCCUGGCCUCCACAUGUGGUGGCAGCGCCUUCAUGCUGUUCAU -----.......(((.(((((.........-...-...(((((((((((((((........))).....))))))))))))......(((((......)))))..)))))))).(((((((......)))))))... ( -41.00, z-score = 0.23, R) >droYak2.chr3R 21577047 130 + 28832112 -----AAAAAUCCCAUUGCGUGUUUUAGUC-UAU-CGACAUGUCCGCCAGCUCCUCAGCCUGCGAUUGUUUGGUGGGCGUGCCCACUGGGCCCACCCUAGCCUCCACAUGUGGCGGUAGUGCCUUCAUGCUGUUCAU -----............(((((.......(-(((-((((((((.(((..(((....)))..)))...(((.(((((((.(.......).)))))))..)))....))))))..))))))......)))))....... ( -39.22, z-score = -0.25, R) >droEre2.scaffold_4770 3347186 130 - 17746568 -----GAAAUUCCCAUUGUGUAUUUCAGUC-AAU-CAACAUGUCCGCCAGCUCCUCAGCCUGCGAUUGUUUGGUGGGCGUGCCCACUGGGCCCACCCUGGCCUCCACAUGUGGUGGUAGUGCCUUUAUGCUGUUCAU -----(((((..((((((((.....((((.-...-...(((((((((((((...((.......))..).))))))))))))...))))((((......))))..)))).))))..))(((........))).))).. ( -35.80, z-score = 0.96, R) >droAna3.scaffold_13340 2873293 135 - 23697760 GUACUUUCUUUUUCAUUUUAUAAUUUAGAA-AAU-AAUCAUGUCCGCCAGCCCCUCAGCCUGCGAUUGCUUAGUGGGCGUACCCACAGGACCCACAUUGGCCUCCACCUGCGGUGGCAGUGCUUUCAUGCUGUUCAU ....(((((.................))))-)..-............((((.....(((((((.(((((...(((((....))))).(((.((.....))..)))....))))).)))).))).....))))..... ( -34.73, z-score = -0.85, R) >dp4.chr2 26493849 113 - 30794189 -----------------------CACAGCCUUAU-CAUAAUGGCCACCAGUCCAUCCUCCUGCGAUUGUUUGGUGGGCGUACCUACGGGGCCCACCCUCGCCUCCACAUGCGGCGGCAGCGCCUUCAUGCUGUUCAU -----------------------.(((((.....-......(((...(((((((......)).)))))...(((((((...((....)))))))))...))).........((((....)))).....))))).... ( -37.80, z-score = -0.61, R) >droPer1.super_6 1822538 113 - 6141320 -----------------------CACAGCCUUAU-CAUAAUGGCCACCAGUCCAUCCUCCUGCGAUUGUUUGGUGGGCGUACCCACGGGGCCCACCCUCGCCUCCACCUGCGGCGGCAGCGCCUUCAUGCUGUUCAU -----------------------.(((((.....-.......((((.(((((((......)).)))))..))))((((((......(((.....)))..(((.((......)).))).))))))....))))).... ( -36.80, z-score = -0.16, R) >droWil1.scaffold_181089 6957274 117 - 12369635 -------------------GCAAUUCCCUGCAAU-CAUCAUGUCCACCAGCACAUCAGCCUGUGAUUGUUUGGUGGGCGUGCCCACUGGACCCACCCUAGCAUCCACAUGUGGCGGCAGCGCCUUUAUGCUAUUCAU -------------------(((....(((((...-...((((((((((((.((((((.....))).)))))))))))))))(((((((((..(......)..))))...)))).))))).)......)))....... ( -35.10, z-score = -0.05, R) >droVir3.scaffold_12855 5937461 123 - 10161210 --------------GCUAUUUCCAGUGAUUUAAUACAACAUGUCCACCAGCACAUCAGCCUGUGAUUGUUUGGUGGGCGUGCCCACCGGCCCAACACUCGCUUCUACAUGCGGCGGCAGCGCAUUUAUGCUGUUCAU --------------((.......((((............(((((((((((.((((((.....))).))))))))))))))(((....)))....))))(((........)))))(((((((......)))))))... ( -37.10, z-score = -0.53, R) >droMoj3.scaffold_6540 6238772 120 - 34148556 --------------GCUAUUCACAGCAAUUCGA---AACAUGUCUACCAGCACAUCAGCUUGUGAUUGUUUGGUGGGCGUGCCCACCGGACCAACACUCGCCUCCACAUGCGGCGGCAGCGCAUUCAUGCUAUUCAU --------------(((......))).....((---(.((((.......((..........(((...((((((((((....))))))))))...)))..(((.((......)).))).)).....))))...))).. ( -36.10, z-score = -0.80, R) >droGri2.scaffold_14906 6053975 122 + 14172833 --------------GCCAAUCACAGUGAUUAAA-ACAACAUGUCCACCAGCACAUCAGCCUGUGAUUGUUUGGUAGGCGUGCCCACCGGCCCAACGCUUGCCUCCACAUGCGGCGGAAGCGCAUUUAUGCUGUUUAU --------------(((((((((((.(((....-.......))).....((......)))))))))))...((((((((((((....)))...))))))))).....(((((.(....))))))....))....... ( -37.00, z-score = -0.60, R) >apiMel3.Group1 5223837 117 - 25854376 -----------------GUGAAGAUUAUUGUAAA-UUUAAUUGAUAUUA-CAGAUGAGC-UGUAACUGUCCAUUAAACCCAUCGACUGGACCAACACUGGCAUCAACAUGCGGUGGUUCGUCCUUUAUGCUAUUCAU -----------------((((((...........-((((((.(((((((-(((.....)-))))).)))).))))))......(((.((((((....(.((((....)))).))))))))))))))))......... ( -26.60, z-score = -1.08, R) >consensus ______________AUUGUGUAUUUUAGUC_AAU_CAACAUGUCCACCAGCACAUCAGCCUGCGAUUGUUUGGUGGGCGUGCCCACUGGGCCCACCCUGGCCUCCACAUGCGGCGGCAGCGCCUUCAUGCUGUUCAU ...............................................((((......((((((....((((((((((....))))))))))........(((.(.....).))).)))).))......))))..... (-22.43 = -22.43 + 0.01)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:55:42 2011