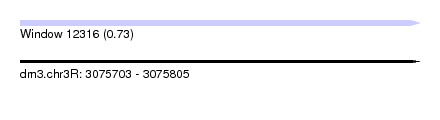
| Sequence ID | dm3.chr3R |
|---|---|
| Location | 3,075,703 – 3,075,805 |
| Length | 102 |
| Max. P | 0.732996 |

| Location | 3,075,703 – 3,075,805 |
|---|---|
| Length | 102 |
| Sequences | 13 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 61.93 |
| Shannon entropy | 0.84002 |
| G+C content | 0.53841 |
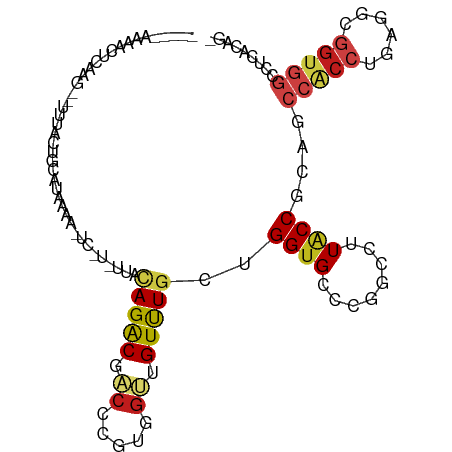
| Mean single sequence MFE | -28.63 |
| Consensus MFE | -9.81 |
| Energy contribution | -9.98 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.05 |
| Structure conservation index | 0.34 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.732996 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
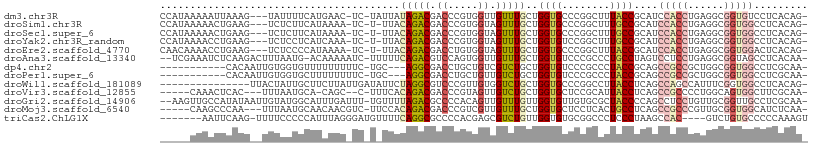
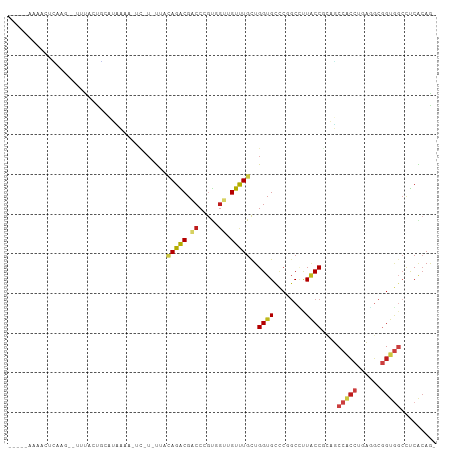
>dm3.chr3R 3075703 102 + 27905053 CCAUAAAAAUUAAAG---UAUUUUCAUGAAC-UC-UAUUAUAGACGACCCGUGGUUGUUUGCUGGUGCCCGGCUUUACCGCAUCCACCUGAGGCGGUGUCCUCACAG- .(((.(((((.....---.))))).)))...-..-......((((((((...))))))))(((((...)))))..((((((.((.....)).)))))).........- ( -23.90, z-score = -0.53, R) >droSim1.chr3R 3119988 101 + 27517382 CCAUAAAAACUGAAG---UCUCUUCAUAAAA-UC-U-UUACAGACGACCCGUGGUAGUUUGCUGGUGCCCGGCUUUGCCGCAUCCACCUGAGGCGGUGGCCUCACAG- .........(((..(---(((..........-..-.-....)))).....(((((((...(((((...))))).))))))).......((((((....)))))))))- ( -28.13, z-score = -0.74, R) >droSec1.super_6 3167849 101 + 4358794 CCAUAAAAACUGAAG---UCUCUUCAUAAAA-UC-U-UUACAGACGACCCGUGGUAGUUUGCUGGUGCCCGGCUUUGCCGCAUCCACCUGAGGCGGUGGCCUCACAG- .........(((..(---(((..........-..-.-....)))).....(((((((...(((((...))))).))))))).......((((((....)))))))))- ( -28.13, z-score = -0.74, R) >droYak2.chr3R_random 824524 101 + 3277457 CCAUAAAACCUGAAG---UCUCCUCAUCAAA-UC-U-UUACAGACGACCCGUGGUAGUUUGCUGGUGUCCGGCUUUGCCGCAUCCACCUGAGGCGGUGGCCUCACAG- .........(((..(---(((..........-..-.-....)))).....(((((((...(((((...))))).))))))).......((((((....)))))))))- ( -27.93, z-score = -0.53, R) >droEre2.scaffold_4770 3339742 101 + 17746568 CAACAAAACCUGAAG---UCUCCCCAUAAAA-UC-U-UUACAGACGACCUGUGGUAGUUUGCUGGUGCCCGGCUUUACCGCAUCCACCUGAGGCGGUGGACUCACAG- .........(((..(---(((..........-..-.-....))))((..((((((((...(((((...))))).))))))))((((((......)))))).)).)))- ( -28.73, z-score = -1.15, R) >droAna3.scaffold_13340 2866179 103 + 23697760 --UCGAAAUCUCAAGACUUUAAUG-ACAAAAAUC-UUUUUCAGACGUCCAGUGGUUGUUUGCUGGUGUCCCGCCCUGCCUAGUCCUCCUGAGGCGGUAGCCUCACAA- --............((((....((-(.(((....-))).)))((((.((((..(....)..))))))))...........))))....((((((....))))))...- ( -25.60, z-score = -0.87, R) >dp4.chr2 26487490 92 + 30794189 -----------CACAAUUGUGGUGUUUUUUUUUC-UGC---AGGCGACCUGCUGUCGUCUGCUGGUGUCCCGCCCUACCGCAGCCGCCGCUGGCGGUGGCCUCGCAA- -----------......(((((((..........-.((---(((((((.....))))))))).((((...)))).)))))))(((((((....))))))).......- ( -40.00, z-score = -2.68, R) >droPer1.super_6 1816192 92 + 6141320 -----------CACAAUUGUGGUGCUUUUUUUUC-UGC---AGGCGACCUGCUGUUGUCUGCUGGUGUCCCGCCCUACCGCAGCCGCCGCUGGCGGUGGCCUCGCAA- -----------......(((((((..........-.((---(((((((.....))))))))).((((...)))).)))))))(((((((....))))))).......- ( -37.80, z-score = -1.73, R) >droWil1.scaffold_181089 6949407 92 + 12369635 ---------------UUACUAUUGCUUCUUAUUGAUAUUCUAGGCGUCCCGUUGUGGUCUGCUGGUGCCCGGCCUUACCUCAGCCAGCCAUUUCGGUGGCCUCACAG- ---------------..........................((((...(((..(((((..(((((.....((.....)))))))..)))))..)))..)))).....- ( -22.80, z-score = 0.25, R) >droVir3.scaffold_12855 5929348 95 + 10161210 -----CAAACUCAC---UUUAAUGCA-CAGC--C-UUUCACAGACGACCCGUAGUUGUCUGCUGGUGCUCCGCAUUACCUCAGCCGCCCCUGGCAGUGGCUUCGCAA- -----.........---..((((((.-.(((--.-..(((((((((((.....)))))))).))).)))..))))))....((((((........))))))......- ( -28.50, z-score = -1.65, R) >droGri2.scaffold_14906 6046601 104 - 14172833 --AAGUUGCCAUAUAAUUGUAUGGCAUUUGAUUU-UGUUUUAGACGCCCCACAGUUGUUUGUUGGUGUUGUGCGCUACCCCAGCCUCCUGUUGCGGUUGCCUCGCAA- --....((((((((....))))))))........-.......((((((..((((....)))).))))))(((.((.(((.((((.....)))).))).))..)))..- ( -29.60, z-score = -1.90, R) >droMoj3.scaffold_6540 6230541 98 + 34148556 -----CAAGCCCAA---UUUAAUGCAACAACGUC-UUCCACAGACGACCCGUCGUUGUUUGCUGGUGCUCCUCACUGCCUCAGCCGCCCGUUGCGGUGGCAUCUCAA- -----..((((((.---......(((((((((((-(.....))))((....)))))).))))))).)))......((((...(((((.....)))))))))......- ( -28.21, z-score = -1.47, R) >triCas2.ChLG1X 7866282 96 + 8109244 -------AAUUCAAG-UUUUCCCCCAUUUAGGGAUGUUUUCAGGCGCCCCACGAGCGUCUGUUGGUGUGCGGCCCUCCCUAAGCCAC----GUCUGUGCCCCCAAAGU -------........-..............(((..((...(((((((..(((.(((....))).))).))(((.........)))..----))))).)).)))..... ( -22.80, z-score = 0.15, R) >consensus _____AAAACUCAAG__UUUACUGCAUAAAA_UC_U_UUACAGACGACCCGUGGUUGUUUGCUGGUGCCCGGCCUUACCGCAGCCACCUGAGGCGGUGGCCUCACAG_ ........................................(((((.((.....)).)))))..((((........))))....(((((......)))))......... ( -9.81 = -9.98 + 0.16)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 23:55:41 2011